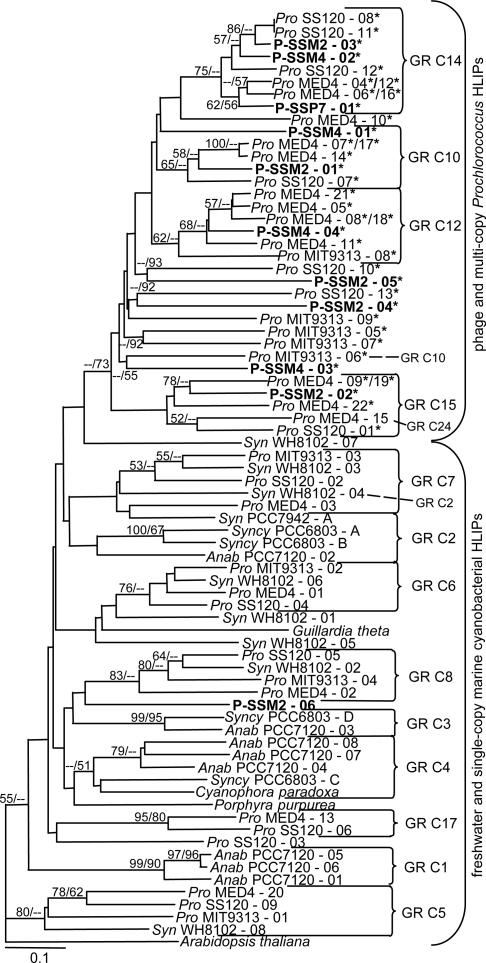
Fig. 3.
Distance tree of HLIPs. Phage HLIPs appear in bold. The tree was generated from 36 amino acids (see Fig. 8), with gaps treated as missing data. generage clusters are indicated to the right of the tree, with cluster designations following ref. 27. Three discrepancies found between generage and distance tree clustering are indicated by the dashed line and their GR cluster designations. Asterisks denote proteins encoding at least six of the nine amino acids of the C-terminal 9-aa consensus sequence. Bootstrap and quartet puzzling values >50% are shown at the nodes for distance and maximum likelihood analyses, respectively. The tree was rooted with the single HLIP from A. thaliana. Abbreviations are as for Fig. 2.