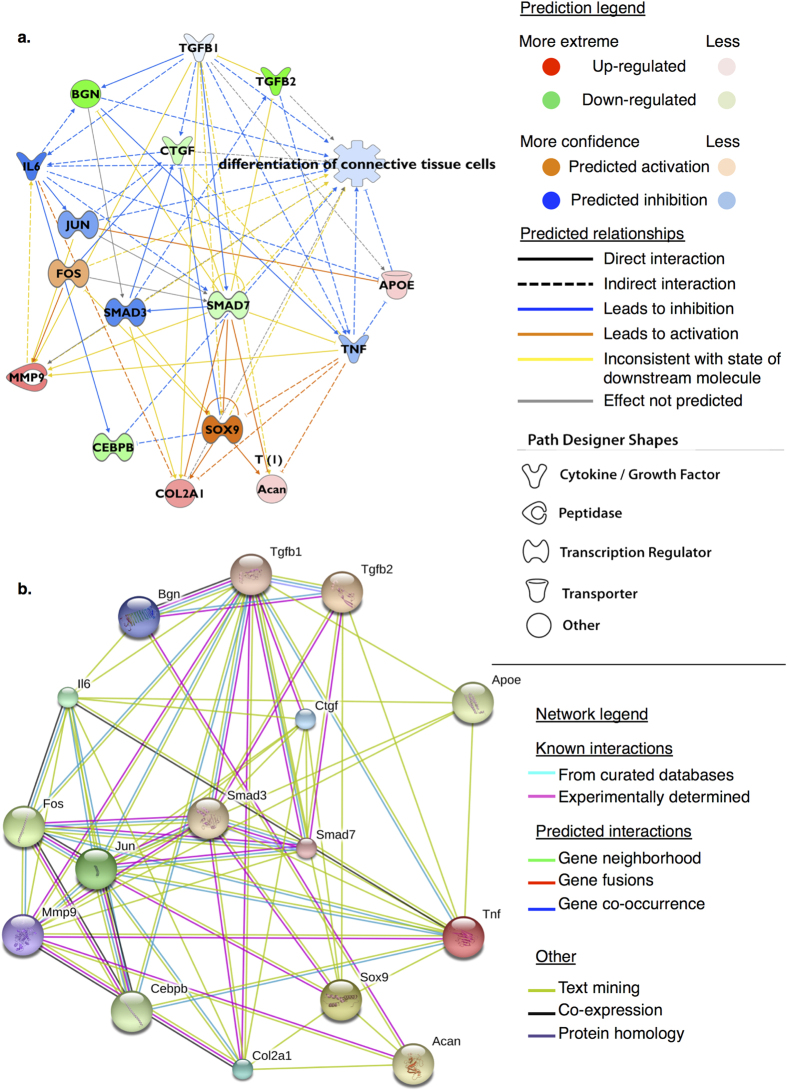
Figure 5.
(a) Hypothetical mechanistic network derived from differentially expressed gene expression profile for cartilage versus dedifferentiated chondrocytes (dedifferentiation transition). Network consists of genes (nodes) connected by lines (edges) indicating a known relationship/interaction in the IPA® Knowledge Base. Genes more highly expressed in cartilage are coloured red; genes showing low expression in cartilage (i.e. higher in monolayer) are coloured green. Figure prediction legend describes the nature of edges joining nodes. Upstream regulators, and intermediate nodes (Jun, Smad3, Il6, Tnf), were predicted to be activated (orange) or inhibited (blue) consistent with the gene expression profile supplied; the direction and nature of the relationship is also indicated. Based upon the observed gene expression profile Tgf-β1 was predicted to be a key upstream regulator (z-score -2.65, overlap p-value 5.41e-26) of 740 differentially expressed genes; the actions of Tgf-β1 were predicted to be inhibited in native cartilage where expression of Smad7, Bgn, and Ctgf were low relative to monolayer chondrocytes at passage five. Functional annotations for ‘differentiation of chondrocytes’ (3.66e-11), and ‘differentiation of connective tissue cells’ (5.6e-21, inhibited) were significantly enriched for this subnetwork and indicated a differentiation process; (b) Using the same elements a protein-protein association network consisting of nodes (proteins) and edges (evidence of associations) indicates a shared function between selected nodes, but not necessarily physical interactions. Sources for evidence of associations are defined in the network legend. Elements determined to influence differentiation status of chondrocytes and tenocytes in culture using IPA® are shown to have significant enrichment for protein-protein interactions (p < 0.0001) indicating that as a group they are biologically connected.