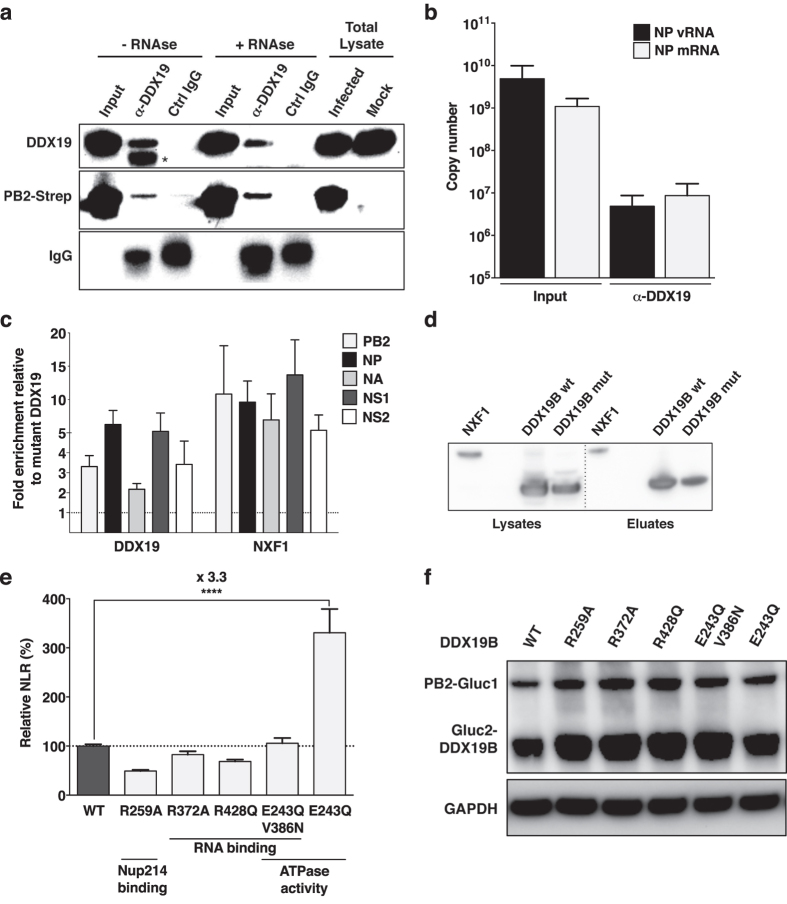
Figure 5. The viral polymerase and viral RNAs interact with DDX19.
(a,b) A549 cells were infected with WSN (5 pfu/cell). At 3.5 hpi, DDX19 proteins were purified in the absence or presence of RNAse, using anti-DDX19 antibodies (α-DDX19) or control immunoglobulins (Ctrl IgG). (a) Inputs and α-DDX19/Ctrl IgG eluates were analyzed by immunoblot to detect DDX19, PB2-Strep and the IgG antibodies (upper, middle and lower panel, respectively). Results representative of two independent experiments are shown. The star indicates a non-specific band. (b) The levels of NP mRNAs and vRNAs co-purified with DDX19 are expressed as the mean ± SEM of four independent experiments. The background level of detection in control IgG eluates was 2.9 × 106 and 3.9 × 105 copies for NP vRNAs and NP mRNAs, respectively. Co-immunoprecipitated to input copy ratios between NP mRNAs and vRNAs differed significantly (multivariate linear model with an interaction term between sample type and RNA type, p = 0.0465). (c,d) HEK-293T cells expressing Strep-tagged wild-type DDX19B, mutant DDX19B or NXF1 were infected with WSN (5 pfu/cell). At 6 hpi, Strep-tagged proteins were purified, and the levels of co-purified mRNAs were determined (c). For wild-type DDX19B and NXF1, co-purified/total mRNAs ratios were normalized to the one obtained with the DDX19 mutant. The data represent the mean ± SEM of three independent experiments. Statistical analysis (multivariate linear model) did not demonstrate significance. (d) Lysates and eluates were analyzed by immunoblot using Strep-Tactin. The dashed line indicates the juxtaposition of non-adjacent lanes from the same immunoblot. (e,f) HEK-293T cells expressing the Gluc2-DDX19B variants were infected with the WSN-PB2-Gluc1 virus at a m.o.i. >1 pfu/cell. At 6 hpi, cells were lysed and normalized luminescence ratios (NLRs) were determined (e). Four independent experiments in triplicate were performed. The results are expressed as the mean percentages ± SEM of luciferase activity relative to the DDX19B wild-type condition. The significance was tested with a Holm-Sidak’s multiple comparisons test using GraphPad Prism software (****p < 0.0001). (f) Lysates were analyzed by immunoblot using anti-Gluc or anti-GAPDH antibodies. Cropped blots are shown in (a,d,f). Corresponding full-length blots are shown in Figures S7, S8 and S9, respectively.