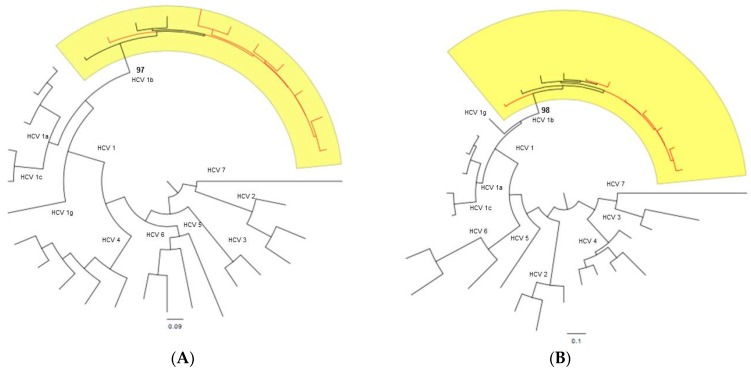
Figure 1.
RAxML phylogenetic trees were estimated using 24 hepatitis C virus (HCV) reference sequences (black) downloaded from Los Alamos HCV Sequence Database and 8 HCV isolates (red) in this study for NS3 (A) and NS5B (B) regions, respectively. The reliability of the phylogenetic clustering was evaluated using bootstrap analysis with 1000 replicates. Bootstrap support values are only shown for the HCV1b clades (yellow area). The scale bars at the bottom of the figure represent genetic distance.