Abstract
Aberrant Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling is crucial to the development of gastric cancer. In this study, we examined the role of STAT3 in the expression and methylation of its targets in gastric cancer patients. Results from RNA sequencing identified an inverse correlation between the expression of STAT3 and GATA6 in 23 pairs of gastric cancer patient samples. We discovered that the expression of GATA6 is epigenetically silenced through promoter methylation in gastric cancer cell lines. Interestingly, the inhibition of STAT3 using a novel STAT3 inhibitor restored the expression of GATA6 and its targets, trefoil factors 1 and 2 (TFF1/2). Moreover, disruption of STAT3 binding to GATA6 promoter by small hairpin RNA restored GATA6 expression in AGS cells. A clinically significant correlation was also observed between the expression of GATA6 and TFF1/2 among tissue samples from 60 gastric cancer patients. Finally, bisulfite pyrosequencing revealed GATA6 methylation in 65% (39/60) of the patients, and those with higher GATA6 methylation tended to have shorter overall survival. In conclusion, we demonstrated that aberrant JAK/STAT signaling suppresses TFF1/2 partially through the epigenetic silencing of GATA6. Therapeutic intervention of STAT3 in reversing the epigenetic status of GATA6 could benefit the treatment of gastric cancer and is worthy of further investigation.
Keywords: epigenetic silencing, gastric cancer, TFF1, TFF2, GATA6, STAT3
1. Introduction
Gastric cancer is the fifth leading cause of cancer death in Taiwan, and infection with Helicobacter pylori is a major risk factor for gastric carcinoma [1]. Previous studies have shown that cytotoxin-associated gene A (CagA) positive H. pylori presents a greater risk of inducing gastric carcinogenesis than does CagA negative H. pylori. Studies have also shown that gastric epithelial cells infected with CagA positive H. pylori can activate the signal transducer and activator of transcription 3 (STAT3) and corresponding JAK/STAT signaling [2]. However, the role that aberrant JAK/STAT signaling plays in epigenetic alteration of its downstream targets has yet to be fully elucidated.
Trefoil factors (TFFs), which are secreted by epithelial cells to interact with glycosylated molecules on mucosal surfaces, play an important role in protecting the stomach. Indeed, several studies have shown that TFFs can protect the epithelia against mucosal damage [3,4]. The expression of TFFs stimulates cellular motility as a means of promoting mucosal defense and wound healing [5]. Two out of three TFFs found in the human stomach (TFF1 and TFF2 but not TFF3), have been identified as tumor suppressors [6]. Thus, TFF1 and TFF2 (TFF1/2) are frequently downregulated in gastric cancer.
GATA6 recognizes the motif (A/T)GATA(A/G) and regulates gene expression, including TFF1/2. In humans, GATA1/2/3 are expressed in hematopoietic cells, whereas GATA4/5/6 are expressed in the cardiovascular system and endoderm-derived tissue, including the liver, lung, and pancreas [7]. GATA6 has been shown to regulate a gastric parietal cell-specific gene that encodes H+/K+-ATPase [8]. Al-azzeh et al. previously found that GATA6 can also activate the expression of TFF1/2 in the stomach [9]. We therefore hypothesized that the downregulation of GATA6 could lead to the downregulation of TFF1/2 in gastric cancer.
In this study, we found that the expression level of GATA6 is inversely correlated with that of STAT3 in gastric cancer patients. A positive correlation was also observed between the expression of GATA6 and TFF1/2. The role of JAK/STAT3 signaling in the suppression of TFF1/2 through epigenetic silencing of GATA6 was also explored in this study.
2. Results
2.1. Inverse Relationship between Expression of STAT3 and GATA6 in Gastric Cancer
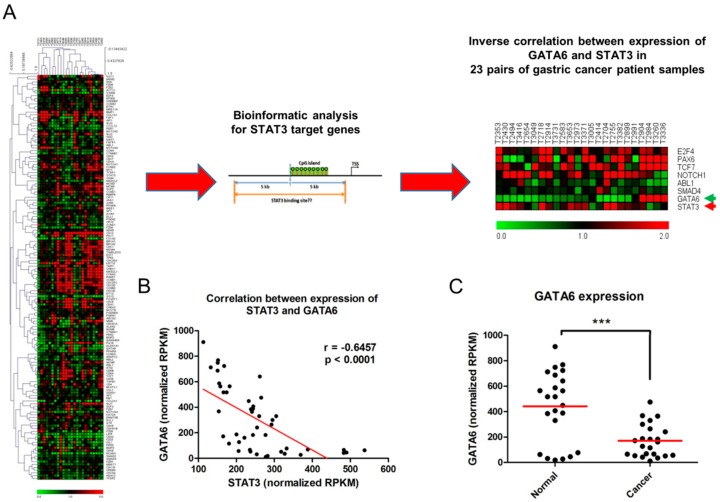
To identify STAT3 targets that are differentially expressed through the activation of STAT3 in gastric cancer, we performed RNA sequencing (RNA-Seq) by using 23 pairs of gastric cancer patient samples (Figure 1A). We further performed bioinformatic analysis to identify genome-wide CpG island promoters containing STAT3-binding sites and to determine the differentially-expressed genes that are regulated by STAT3 (Figure 1A and Figure S1). Combined results from expression arrays and bioinformatic analyses revealed an inverse correlation between the expression of STAT3 and GATA6 in those patient samples (Figure 1B). Interestingly, adjacent normal tissues showed a significantly higher expression of GATA6 than that of cancer tissues (Figure 1C).
Figure 1.
Inverse correlation between the expression of signal transducer and activator of transcription 3 (STAT3) and GATA6 in gastric cancer: (A) Heatmap showing the expression profile for 23 pairs of gastric cancer patients investigated in the RNA-Seq experiment (Please refer to Figure S1 for a higher resolution version of the heatmap). Bioinformatic analysis was used to identify potential STAT3 targets by filtering genes with at least one STAT3 binding site within 5 kb of the promoter CpG island. Integration of RNA-Seq and bioinformatic analyses identified several genes, including GATA6, which were correlated with the expression of STAT3; (B) Scatter plot illustrates the inverse correlation between the expression of STAT3 and GATA6 among 23 pairs of gastric cancer patients; (C) Dot plot shows a significantly higher expression of GATA6 in adjacent normal tissues than that of cancer tissues (*** p < 0.001). Red bar indicates median. TSS: transcription start site; RPKM: reads per kilobase per million.
2.2. Epigenetic Silencing of GATA6 by Aberrant JAK/STAT Signal Suppresses TFF1/2
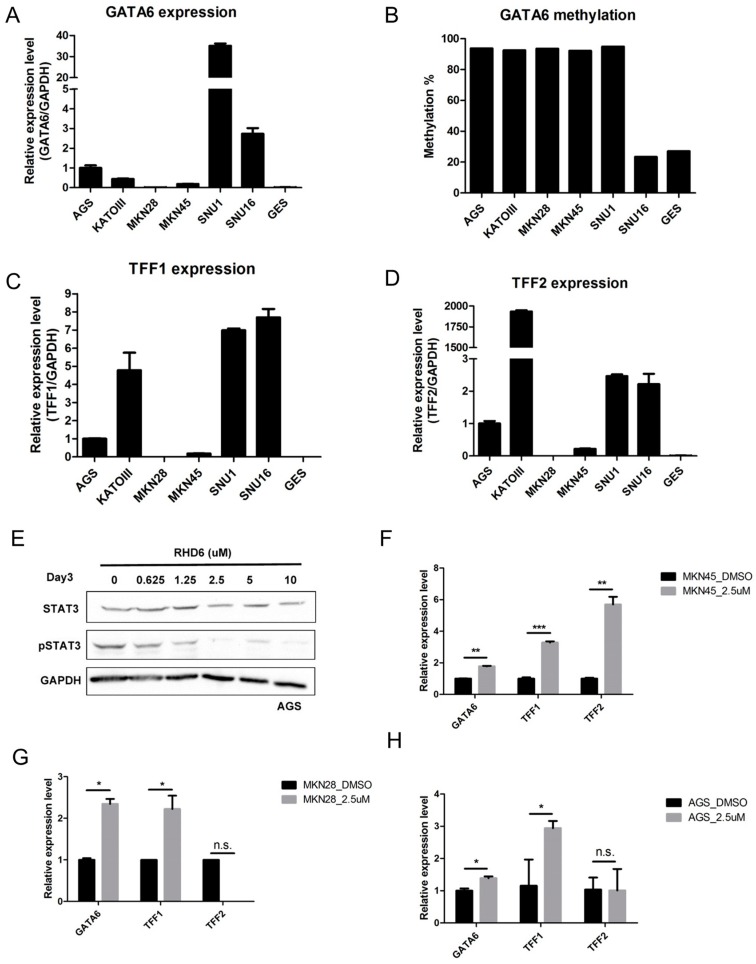
Previous researchers have reported that promoter methylation of GATA6 was observed in human cancer [10,11]. To determine whether this epigenetic event also occurs in gastric cancer, we examined expression and methylation status in various gastric cancer cell lines. The expression of GATA6 (Figure 2A,B) was correlated with promoter methylation in all gastric cancer cell lines except for SNU1 and GES cells. Given that GATA6 is a transcription factor of the gastroprotective trefoil genes TFF1/2 [9], we therefore examined the expression of TFF1/2 in gastric cancer cell lines. As expected, the expression of TFF1/2 presented a positive correlation with that of GATA6 in most of the cells.
Figure 2.
Epigenetic silencing of GATA6 through the activation of STAT3 suppresses expression of trefoil factors (TFF) 1 and 2 in gastric cancer: (A) Relative expression levels of GATA6 in immortalized gastric epithelial cells (GES) and gastric cancer cell lines as determined by RT-PCR; (B) Methylation level of the GATA6 promoter in gastric cancer cell lines, as determined by bisulfite pyrosequencing. Relative expression levels of (C) TFF1 and (D) TFF2 in the same gastric cancer cell lines, as determined by RT-PCR; (E) Western blot analysis showing the protein levels of STAT3 and phosphorylated STAT3 (pSTAT3) in AGS gastric cancer cells treated with various quantities of STAT3 inhibitor rhodium(III) complex 6 (RHD6). The protein level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. Relative expression levels of GATA6, TFF1, and TFF2 in (F) MKN45, (G) MKN28, and (H) AGS gastric cancer cells treated with dimethyl sulfoxide (DMSO) or 2.5 μm of RHD6. (*** p < 0.001; ** p < 0.01; * p < 0.05). Each bar represents mean ± standard deviation (SD) of duplicate experiments. n.s.: not significant.
The above experiments demonstrate that GATA6 is epigenetically silenced in gastric cancer. In our previous study on ovarian cancer, we demonstrated that aberrant signaling may lead to the epigenetic silencing of its downstream targets [12,13]. In the current study, gastric cancer cells were then treated with rhodium(III) complex 6 (RHD6) [14], a novel STAT3 inhibitor, to determine whether the suppression of STAT3 would derepress GATA6 and TFF1/2. We found that RHD6 treatment suppressed STAT3 phosphorylation but not total STAT3 in AGS cells (Figure 2E). Importantly, treatment with 2.5 µm of RHD6 restored the expression of GATA6 and TFF1/2 in AGS, MKN28, and MKN45 gastric cancer cells (Figure 2F–H). Similar results can be found in AGS cells treated with the JAK inhibitor, AG490 (Figure S2).
2.3. STAT3 Binds to the GATA6 Promoter
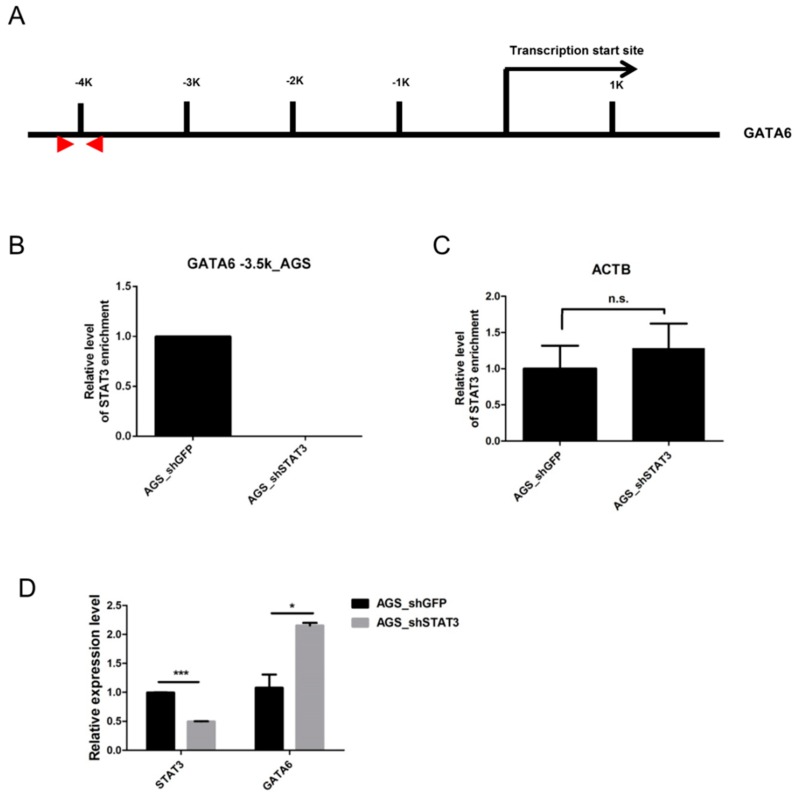
To determine whether STAT3 affects the expression of GATA6 directly, we examined the binding status of STAT3 in AGS cells showing constitutive activation of STAT3 [15,16]. The binding of STAT3 can be observed at the putative STAT3 binding site upstream of the GATA6 promoter (Figure 3A). The binding at the GATA6 promoter (Figure 3B) but not β-actin (ACTB, negative control, Figure 3C) was disrupted upon STAT3 depletion in AGS cells. Importantly, disruption of STAT3 restored GATA6 expression in AGS cells (Figure 3D).
Figure 3.
Binding of STAT3 represses GATA6 expression in AGS gastric cancer cells: Using lentiviral small hairpin RNA (shRNA), chromatin immunoprecipitation (ChIP)-PCR experiments were performed to examine the relative binding of STAT3 to the GATA6 promoter in AGS cells and cells with depleted STAT3. (A) The genomic structure of the GATA6 promoter and the location of the ChIP-PCR primer (red arrow heads) targeting the putative STAT3 binding site at the upstream promoter region. STAT3 binding was significantly enriched in the (B) GATA6 promoter but not (C) β-actin (ACTB) promoter (negative control) in AGS cells; (D) Depletion of STAT3 significantly restored GATA6 expression in AGS cells. *** p < 0.005, * p < 0.05. n.s.: not significant.
2.4. Expression of GATA6 Correlates with TFF1/2 in Gastric Cancer Patients
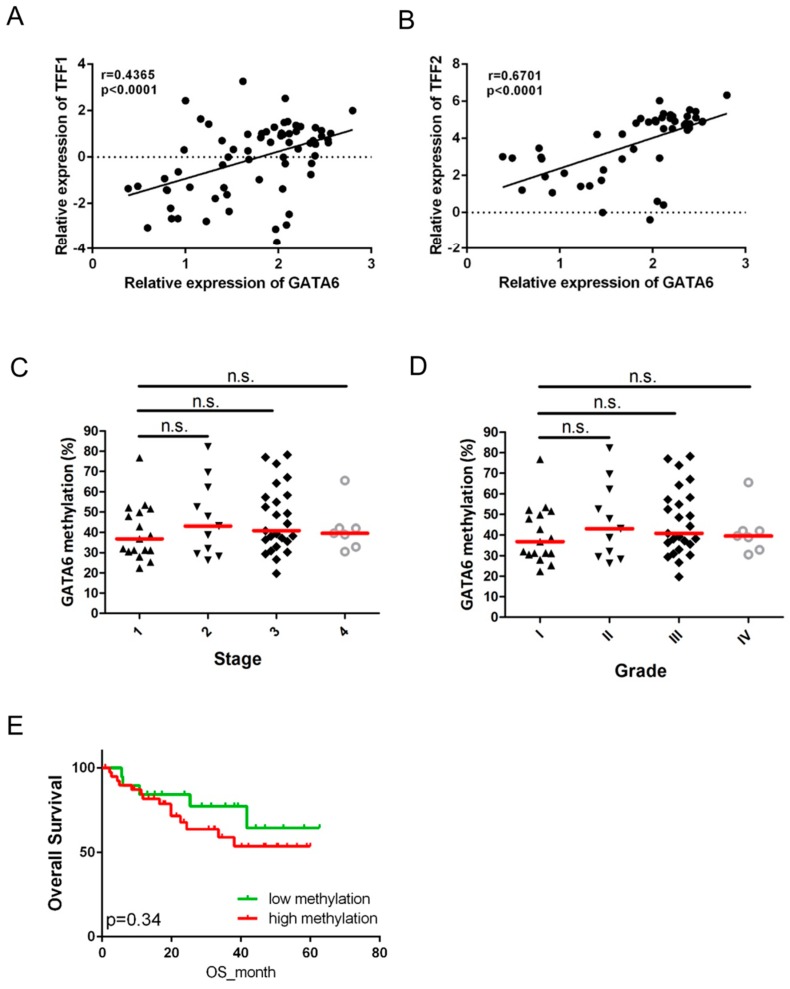
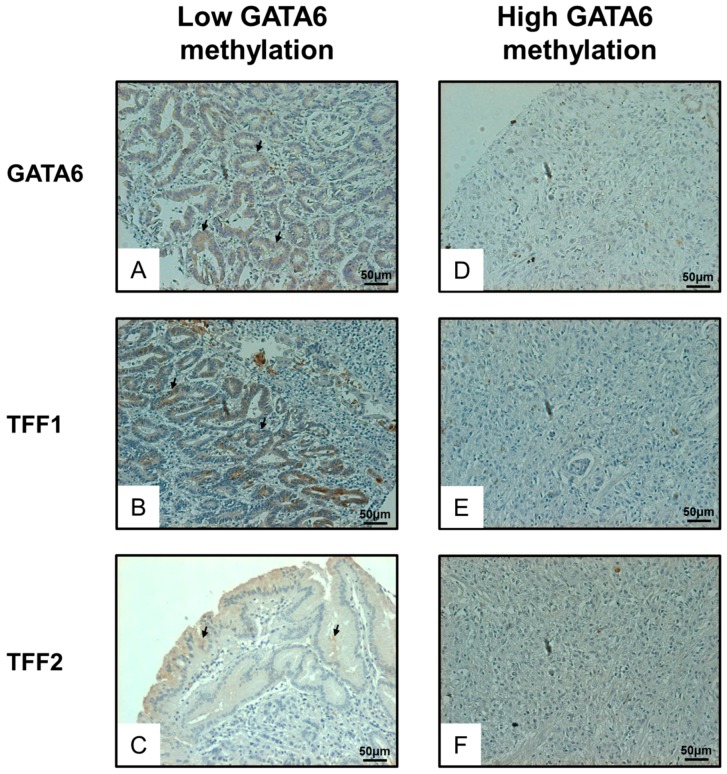
A significantly positive correlation was observed between the expression of GATA6 and TFF1/2 in the full set of patients with gastric cancer (n = 60, Table 2, Figure 4A,B). As expected, samples with low GATA6 methylation but not high GATA6 expression demonstrated a higher protein expression of GATA6 as well as TFF1/2 (Figure 5). We also examined the clinical significance of GATA6 methylation (p < 0.0001, Figure 4C,D). Despite the fact that GATA6 methylation was not significantly associated with any clinical parameters (Table 1), patients with higher GATA6 methylation tended to have shorter overall survival than did those with lower GATA6 methylation (Figure 4E).
Table 2.
Summary of clinico-pathological data for the gastric cancer patients in this study.
| RNA-Seq (n = 23) | Full Set (n = 60) | |
|---|---|---|
| Age | ||
| Median | 68.5 | 68.5 |
| Range | 47–87 | 47–87 |
| Stage | ||
| I | 3 | 17 |
| II | 5 | 11 |
| III | 11 | 25 |
| IV | 4 | 7 |
| Grade | ||
| I | 3 | 17 |
| II | 5 | 11 |
| III | 11 | 25 |
| IV | 4 | 7 |
| Median survival | 23.53 | 27.1 |
| H. pylori | ||
| positive | 16 | 44 |
| negative | 8 | 16 |
Figure 4.
Promoter methylation of GATA6 is frequently observed in gastric cancer patients. Scatter plots showing the correlation between the expression of GATA6 and (A) TFF1 and (B) TFF2 among 60 gastric cancer patients. A significant correlation was observed between GATA6 and TFF1/2. The dot plot shows the relationship between methylation level of the GATA6 promoter and (C) tumor stage or (D) tumor grade among 60 patients with gastric cancer; (E) Results from a Kaplan–Meier analysis which show that gastric cancer patients with higher GATA6 methylation (red line, >35% methylation, see Materials and Methods) tended to have shorter overall survival than did patients with lower GATA6 methylation (green line). OS: overall survival. n.s.: not significant.
Figure 5.
Immunohistochemistry examination of GATA6, TFF1 and TFF2 in gastric cancer patient samples: Samples with low GATA6 methylation (left panel) have higher expression of GATA6 (A) as well as TFF1 (B) and TFF2 (C) as indicated by brown color (arrow). In contrast, samples with high GATA6 methylation (right panel) have lower expression of GATA6 (D), TFF1 (E), and TFF2 (F). Magnification, 100×.
Table 1.
Relationship between clinical parameters and GATA6 methylation among the 60 patients with gastric cancer.
| GATA6 Methylation % 1 (n) | p | |
|---|---|---|
| Age | 0.163 | |
| ≥60 | 50.36 ± 5.48% (10) | |
| <60 | 42.82 ± 2.12% (50) | |
| Sex | 0.873 | |
| Male | 43.87 ± 2.18% (43) | |
| Female | 44.59 ± 4.55% (17) | |
| Stage | 0.497 | |
| I–II | 42.60 ± 2.99% (28) | |
| III–IV | 45.36 ± 2.72% (32) | |
| Grade 2 | 0.497 | |
| Low | 42.60 ± 2.99% (28) | |
| High | 45.36 ± 2.72% (32) |
1 Mean methylation % ± standard deviation (SD); 2 Pathological grade, low: G1–2; high: G3–4.
3. Discussion
The activation of JAK/STAT signaling plays a crucial role in gastric carcinogenesis [2,17]; however, the role of JAK/STAT signaling in the epigenetic silencing of its targets has not been fully explored. In the current study, we examined the expression profile of STAT3 and its putative targets. Our results demonstrate that there is an inverse relationship between the expression of STAT3 and GATA6 in gastric cancer patients. Cell line study also suggests that the expression of GATA6 may be epigenetically controlled by promoter methylation. Importantly, inhibition of JAK/STAT3 signaling by the novel STAT3 inhibitor derepressed GATA6 expression in several gastric cancer cell lines suggesting that STAT3 is a transcription repressor for GATA6. It is noteworthy that GES cells with a low level of GATA6 promoter methylation did not express GATA6. On the contrary, SNU1 cells showing high levels of GATA6 methylation also showed a high level of GATA6 expression (Figure 2A,B). It is thus suggested that other mechanisms such as genetic alteration or histone modifications may be responsible for controlling GATA6 expression in these cells.
In the trefoil peptide family, TFF1 and TFF2 but not TFF3 are important in maintaining the mucosal integrity of the gastrointestinal tract [4]. Previous studies have demonstrated that TFF1/2 are downregulated in gastric cancer, which suggests that they may be tumor suppressors. In the current study, we observed a positive correlation between the expression of GATA6 and TFF1/2 in patients with gastric cancer and in gastric cancer cell lines. Importantly, the inhibition of JAK/STAT signaling by the STAT3 inhibitor restored GATA6 expression as well as the expression of TFF1/2 in cancer cell lines.
A previous study demonstrated that GATA6 is an important transcription factor in the transactivation of TFF1/2 in gastric cancer [9]; however, the current study may provide mechanistic evidence to explain how TFF1 and TFF2 are downregulated by epigenetic silencing of GATA6 through the activation of JAK/STAT signaling in gastric cancer. It is also noted that the expression of TFF1/2 did not correlate well with that of GATA6 in KATOIII cells (Figure 2A–D). The role of factors other than GATA6 in the transcriptional regulation of TFF1/2 cannot be excluded.
This study also demonstrated that GATA6 is epigenetically silenced by promoter methylation in patients with gastric cancer. The methylation of the GATA family has previously been described in several types of human cancer [18,19,20]. Specifically, the methylation of GATA4 and GATA5 has been frequently observed in gastric cancer, which suggests that these genes may be tumor suppressors [21]. We are therefore the first to report that GATA6 can be epigenetically silenced by promoter methylation in gastric cancer. Although the methylation of GATA6 did not correlate with any of the clinical parameters, frequent methylation of GATA6 was observed in our sample cohort, and patients with higher GATA6 methylation tended to have shorter overall survival than those with lower GATA6 methylation. These results suggest that GATA6 may act as a tumor suppressor in gastric cancer and methylation of GATA6 may be an early event in gastric carcinogenesis. Methylation of GATA6 may be able to serve as an early diagnostic marker in gastric cancer. However, further functional and clinical experiments will have to be conducted to test the supposition.
In conclusion, aberrant JAK/STAT signaling may be involved in the epigenetic suppression of GATA6 expression via promoter hypermethylation. The suppression of GATA6 may further downregulate TFF1/2 in gastric cancer. Pharmacological inhibition of STAT3 may restore the expression of GATA6 and the gastric protective factor TFF1/2. Determining the potential role of the STAT3 inhibitor in the treatment of gastric cancer is worthy of further investigation.
4. Materials and Methods
4.1. Patient Sampling
Sixty surgical tissue samples of gastric cancer were obtained from the Tissue Bank, Department of Medical Research, Chang Gung Memorial Hospital, Chiayi, Taiwan (Table 2). In this cohort of patients with gastric cancer, the median age at the time of diagnosis was 68.5 years (range: 47–87 years). Out of these samples, 23 tumor-adjacent normal pairs were used to perform RNA-Seq (Table 2). All assessments conducted in this study were approved by the Institutional Review Board (IRB) of the Chang Gung Memorial Hospital, Chiayi, Taiwan (IRB no.: 102-1993B, approval date: 2013/08/08).
4.2. Cell Culture
The gastric cancer cell lines AGS, KatoIII, MKM28, MKN45, SNU1, SNU16 and an immortalized gastric epithelial cell line, GES (a kind gift from Dr. Jun Yu, The Chinese University of Hong Kong, Hong Kong), were propagated in Roswell Park Memorial Institute medium (RPMI)-1640 (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum and incubated at 37 °C under a humidified atmosphere containing 5% CO2. Cells were treated with various quantities (control, 0.625 μM, 1.25 μM, 2.5 μM, 5 μM, and 10 μM) of novel STAT3 inhibitor RHD6 [14] for 3 days or 20 μm of AG490 (Sigma, St. Louis, MO, USA) for 24 h. The cells were then harvested for protein and RNA extraction.
4.3. DNA Extraction, RNA Extraction, and Quantitative Reverse Transcription-PCR
DNA was extracted using the Tissue & Cell Genomic DNA Purification Kit (Genemark, Taipei, Taiwan). The DNA was eluted in 50 µL distilled water and stored at −20 °C until use. Total RNA from cell lines was extracted using Trizol (Invitrogen) in accordance with the manufacturer’s protocol. Briefly, 1 µg of total RNA was treated with DNase I (amplification grade, Invitrogen) prior to performing first-strand cDNA synthesis using reverse transcriptase (Superscript II RT, Invitrogen). Real-time PCR reactions were conducted using the ABI Step-One real-time PCR system (Applied Biosystems, Foster City, CA, USA) with specific primers (Table 3). The relative expression was calculated using the comparative Ct method.
Table 3.
Primer sequences used in this study.
| Sequence 5′–3′ | |
|---|---|
| RT Primer | |
| STAT3 forward | ACTTTCACTTGGGTGGAGAAGGACAT |
| STAT3 reverse | CTGCTGCTTTGTGTATGGTTCCA |
| GAPDH forward | CCCCTTCATTGACCTCAACTACAT |
| GAPDH reverse | CGCTCCTGGAAGATGGTGA |
| GATA6 forward | AGCCGGCCCCTCATCAAGCCGCAGAA |
| GATA6 reverse | AGTTGGCACAGGACAATCCAAGCC |
| TFF1 forward | CACCATGGAGAACAAGGTGA |
| TFF1 reverse | TGACACCAGGAAAACCACAA |
| TFF2 forward | ATGGATGCTGTTTCGACTCC |
| TFF2 reverse | AGAAGCAGCACTTCCGAGAG |
| ChIP-PCR | |
| GATA6 forward | CGATCACGGAAAGACACCTT |
| GATA6 reverse | CCAATGACCGACGAAAGATT |
| ACTB forward | TGCGTGACATTAAGGAGAAG |
| ACTB reverse | GCTCGTAGCTCTTCTCCA |
| Pyrosequencing | |
| GATA6 forward | GGGTGGGGGAGATTTGTAAG |
| GATA6 reverse | AGCTGGACATCACCTCCCACAACGAAACCTT |
| CTCCCTTATACCATATTTCTTCC | |
| GATA6 sequencing | GAGATTTAAATTTAAAGAAAATTAT |
| UB03 biotin primer | AGCTGGACATCACCTCCCACAACG |
RT: reverse transcription; STAT3: signal transducer and activator of transcription 3; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; TFF: trefoil factor; ChIP-PCR: chromatin immunoprecipitation-PCR; ACTB: β-actin.
4.4. Bisulfite Conversion and Pyrosequencing
Bisulfite pyrosequencing was performed as previously described [13]. Briefly, 0.5 µg of genomic DNA was bisulfite-modified using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) in accordance with the manufacturer’s protocol. The bisulfite-modified DNA was subjected to PCR amplification using a tailed reverse primer in combination with a biotin-labeled universal primer. PCR and sequencing primers were designed using PyroMark Assay Design 2.0 software (Qiagen GmbH, Hilden, Germany). GATA6 transcription start site (−3617 to −3352) was PCR amplified using specific primers (Table 3) in a 25 μL reaction using Invitrogen Platinum™ DNA Polymerase (Invitrogen). Prior to pyrosequencing, 1.5 μL of each PCR reaction was analyzed on 1% agarose gel. Pyrosequencing was performed on the PyroMark Q24 (Qiagen) using Pyro Gold Reagents (Qiagen) in accordance with the manufacturer’s protocol. We measured the methylation level of eleven CpG sites located from −3524 to −3439 bp. The methylation percentage of each cytosine was determined by dividing the fluorescence intensity of cytosines with the sum of the fluorescence intensity for cytosines and thymines at each CpG site. In vitro methylated DNA (Merck Millipore, Billerica, MA, USA) was included as a positive control for pyrosequencing.
4.5. RNA-Seq
Total RNA from 23 tumor-normal pairs of gastric cancer tissues (Table 2) was extracted using TRIzol (Invitrogen) in accordance with manufacturer's instructions. RNA-Seq was then performed using Illumina MiSeq (Illumina, San Diego, CA, USA) at the sequencing core of the National Chung Cheng University (Chiayi, Taiwan).
4.6. Protein Extraction and Western Blotting
Cells were lysed with 100 μL of PRO-PREP Protein Extraction Solution (iNtRON Biotechnology, Seongnam, Korea) and protein concentrations determined by Bio-Rad Protein Assay kit (Bio-Rad, Hercules, CA, USA), according to the manufacturer’s protocol. Samples and pre-stained protein markers were electrophoresed through 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and then transferred to polyvinylidene fluoride (PVDF) membranes, using the Mini Trans-Blot Electrophoretic Transfer Cell system (Bio-Rad). The membrane was then incubated overnight at 4 °C with primary antibodies, rabbit anti-pSTAT3 (1:1000, Cell Signaling, Beverly, MA, USA), mouse anti-STAT3 (1:1000, Cell Signaling), rabbit anti-GATA6 (1:1000, Abcam, Cambridge, UK), rabbit anti-TFF1 (1:1000, Abcam), mouse anti-TFF2 (1:1000, Abcam) or mouse anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies (1:1000, Thermo Fisher Scientific, Rockford, IL, USA), as diluted in 1× phosphate-buffered saline with Tween (PBST). The membranes were incubated at room temperature for 1 h, with secondary antibodies (Thermo Fisher, anti-mouse, 1:1000 or anti-rabbit 1:1000, diluted in 1× PBST). Proteins were detected by an enhanced chemiluminescence horseradish peroxidase (HRP) substrate detection kit (Merck Millipore) and MiniChemiTM Chemiluminescence imaging system (Sage Creation, Beijing, China).
4.7. Immunohistochemical Analysis
Paraffin-embedded gastric cancer tissue sections of selected cases from the above-mentioned patients were dewaxed in xylene and rehydrated in alcohol. Antigen retrieval was performed by heating each section at 100 °C for 25 min in 1× NovocastraTM Epitope Retrieval Solution (pH = 9.0, Leica Biosystems, Newcastle, UK). After 5 min rinses in phosphate-buffered saline (PBS), we followed the protocol of NovolinkTM MaxPolymer Detection System (Leica, Wetzlar, Germany) to stain patient samples, with rabbit anti-GATA6 (1:50, Abcam), rabbit anti-TFF1 (1:50, Abcam), or mouse anti-TFF2 (1:50, Abcam) at 4 °C, overnight, followed by secondary antibody containing post primary reagent and HRP-linked polymer. Slides were finally counterstained with hematoxylin.
4.8. Statistical Analysis
Overall survival (OS) was assessed using Kaplan–Meier analysis and the log-rank test. Overall survival was defined as the time period between the day of diagnosis and death. A DNA methylation level of 35% was used as the cut-off, because this is the median level of methylation in gastric cancer samples. An unpaired t-test was also used to compare parameters in the various groups. All statistical calculations were performed using the SPSS statistical package (version 16.0) for Windows (IBM, Chicago, IL, USA). In this study, a p value of <0.05 was considered statistically significant.
Acknowledgments
The authors would like to thank the Tissue Bank, Department of Medical Research, Chang Gung Memorial Hospital at Chiayi for providing all the patient samples. This study was supported by research grants from Chang Gung Memorial Hospital, Taiwan (CORPG6D0011~13 and CORPG6D0031~33).
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/17/9/1467/s1.
Author Contributions
Cheng-Shyong Wu, Kuo-Liang Wei, Jian-Liang Chou, Chung-Kuang Lu, Ching-Chuan Hsieh, Jora M. J. Lin, Yi-Fang Deng, and Wan-Ting Hsu performed the experiments. Cheng-Shyong Wu, Kuo-Liang Wei, and Ching-Chuan Hsieh collected samples from patients. Hui-Min David Wang, Chung-Hang Leung, and Dik-Lung Ma provided the STAT3 inhibitor. Chin Li and Michael W. Y. Chan designed the experiments. Jian-Liang Chou and Michael W. Y. Chan wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pandey R., Misra V., Misra S.P., Dwivedi M., Kumar A., Tiwari B.K. Helicobacter pylori and gastric cancer. Asian Pac. J. Cancer Prev. 2010;11:583–588. [PubMed] [Google Scholar]
- 2.Lee I.O., Kim J.H., Choi Y.J., Pillinger M.H., Kim S.Y., Blaser M.J., Lee Y.C. Helicobacter pylori CagA phosphorylation status determines the gp130-activated SHP2/ERK and JAK/STAT signal transduction pathways in gastric epithelial cells. J. Biol. Chem. 2010;285:16042–16050. doi: 10.1074/jbc.M110.111054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre O., Chenard M.P., Masson R., Linares J., Dierich A., LeMeur M., Wendling C., Tomasetto C., Chambon P., Rio M.C. Gastric mucosa abnormalities and tumorigenesis in mice lacking the pS2 trefoil protein. Science. 1996;274:259–262. doi: 10.1126/science.274.5285.259. [DOI] [PubMed] [Google Scholar]
- 4.Mashimo H., Wu D.C., Podolsky D.K., Fishman M.C. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science. 1996;274:262–265. doi: 10.1126/science.274.5285.262. [DOI] [PubMed] [Google Scholar]
- 5.Kindon H., Pothoulakis C., Thim L., Lynch-Devaney K., Podolsky D.K. Trefoil peptide protection of intestinal epithelial barrier function: Cooperative interaction with mucin glycoprotein. Gastroenterology. 1995;109:516–523. doi: 10.1016/0016-5085(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 6.Katoh M. Trefoil factors and human gastric cancer (review) Int. J. Mol. Med. 2003;12:3–9. doi: 10.3892/ijmm.12.1.3. [DOI] [PubMed] [Google Scholar]
- 7.Viger R.S., Guittot S.M., Anttonen M., Wilson D.B., Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol. Endocrinol. 2008;22:781–798. doi: 10.1210/me.2007-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishi T., Kubo K., Hasebe M., Maeda M., Futai M. Transcriptional activation of H+/K+-ATPase genes by gastric GATA binding proteins. J. Biochem. 1997;121:922–929. doi: 10.1093/oxfordjournals.jbchem.a021674. [DOI] [PubMed] [Google Scholar]
- 9.Al-azzeh E.D., Fegert P., Blin N., Gott P. Transcription factor GATA-6 activates expression of gastroprotective trefoil genes TFF1 and TFF2. Biochim. Biophys. Acta. 2000;1490:324–332. doi: 10.1016/S0167-4781(00)00013-0. [DOI] [PubMed] [Google Scholar]
- 10.Martinez R., Martin-Subero J.I., Rohde V., Kirsch M., Alaminos M., Fernandez A.F., Ropero S., Schackert G., Esteller M. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–264. doi: 10.4161/epi.9130. [DOI] [PubMed] [Google Scholar]
- 11.Guo M., Akiyama Y., House M.G., Hooker C.M., Heath E., Gabrielson E., Yang S.C., Han Y., Baylin S.B., Herman J.G., et al. Hypermethylation of the GATA genes in lung cancer. Clin. Cancer Res. 2004;10:7917–7924. doi: 10.1158/1078-0432.CCR-04-1140. [DOI] [PubMed] [Google Scholar]
- 12.Chou J.L., Su H.Y., Chen L.Y., Liao Y.P., Hartman-Frey C., Lai Y.H., Yang H.W., Deatherage D.E., Kuo C.T., Huang Y.W., et al. Promoter hypermethylation of FBXO32, a novel TGF-β/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab. Investig. 2010;90:414–425. doi: 10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh K.T., Chen T.H., Yang H.W., Chou J.L., Chen L.Y., Yeh C.M., Chen Y.H., Lin R.I., Su H.Y., Chen G.C., et al. Aberrant TGFβ/SMAD4 signaling contributes to epigenetic silencing of a putative tumor suppressor, RunX1T1, in ovarian cancer. Epigenetics. 2011;6:727–739. doi: 10.4161/epi.6.6.15856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma D.L., Liu L.J., Leung K.H., Chen Y.T., Zhong H.J., Chan D.S., Wang H.M., Leung C.H. Antagonizing STAT3 dimerization with a rhodium(III) complex. Angew. Chem. Int. Ed. Engl. 2014;53:9178–9182. doi: 10.1002/anie.201404686. [DOI] [PubMed] [Google Scholar]
- 15.To K.F., Chan M.W., Leung W.K., Ng E.K., Yu J., Bai A.H., Lo A.W., Chu S.H., Tong J.H., Lo K.W., et al. Constitutional activation of IL-6-mediated JAK/STAT pathway through hypermethylation of SOCS-1 in human gastric cancer cell line. Br. J. Cancer. 2004;91:1335–1341. doi: 10.1038/sj.bjc.6602133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh C.M., Chang L.Y., Lin S.H., Chou J.L., Hsieh H.Y., Zeng L.H., Chuang S.Y., Wang H.W., Dittner C., Lin C.Y., et al. Epigenetic silencing of the NR4A3 tumor suppressor, by aberrant JAK/STAT signaling, predicts prognosis in gastric cancer. Sci. Rep. 2016;6:31690. doi: 10.1038/srep31690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J., Dong Y., Kang W., Go M.Y., Tong J.H., Ng E.K., Chiu P.W., Cheng A.S., To K.F., Sung J.J., et al. Helicobacter pylori-induced STAT3 activation and signalling network in gastric cancer. Oncoscience. 2014;1:468–475. doi: 10.18632/oncoscience.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao Y.F., Fang F., Hu S.Y., Lu J., Cao L., Zhao W.L., Xiao P.F., Li Z.H., Wang N.N., Xu L.X., et al. Hypermethylation of the GATA binding protein 4 (GATA4) promoter in Chinese pediatric acute myeloid leukemia. BMC Cancer. 2015;15:1467. doi: 10.1186/s12885-015-1760-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cecener G., Tunca B., Egeli U., Bekar A., Tezcan G., Erturk E., Bayram N., Tolunay S. The promoter hypermethylation status of GATA6, MGMT, and FHIT in glioblastoma. Cell. Mol. Neurobiol. 2012;32:237–244. doi: 10.1007/s10571-011-9753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skiriute D., Vaitkiene P., Saferis V., Asmoniene V., Skauminas K., Deltuva V.P., Tamasauskas A. MGMT, GATA6, CD81, DR4, and CASP8 gene promoter methylation in glioblastoma. BMC Cancer. 2012;12:1467. doi: 10.1186/1471-2407-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama Y., Watkins N., Suzuki H., Jair K.W., van Engeland M., Esteller M., Sakai H., Ren C.Y., Yuasa Y., Herman J.G., et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol. Cell. Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.