ABSTRACT
The use of high-throughput sequencing technologies with the 16S rRNA gene for characterization of bacterial and archaeal communities has become routine. However, the adoption of sequencing methods for eukaryotes has been slow, despite their significance to natural and engineered systems. There are large variations among the target genes used for amplicon sequencing, and for the 18S rRNA gene, there is no consensus on which hypervariable region provides the most suitable representation of diversity. Additionally, it is unclear how much PCR/sequencing bias affects the depiction of community structure using current primers. The present study amplified the V4 and V8-V9 regions from seven microalgal mock communities as well as eukaryotic communities from freshwater, coastal, and wastewater samples to examine the effect of PCR/sequencing bias on community structure and membership. We found that degeneracies on the 3′ end of the current V4-specific primers impact read length and mean relative abundance. Furthermore, the PCR/sequencing error is markedly higher for GC-rich members than for communities with balanced GC content. Importantly, the V4 region failed to reliably capture 2 of the 12 mock community members, and the V8-V9 hypervariable region more accurately represents mean relative abundance and alpha and beta diversity. Overall, the V4 and V8-V9 regions show similar community representations over freshwater, coastal, and wastewater environments, but specific samples show markedly different communities. These results indicate that multiple primer sets may be advantageous for gaining a more complete understanding of community structure and highlight the importance of including mock communities composed of species of interest.
IMPORTANCE The quantification of error associated with community representation by amplicon sequencing is a critical challenge that is often ignored. When target genes are amplified using currently available primers, differential amplification efficiencies result in inaccurate estimates of community structure. The extent to which amplification bias affects community representation and the accuracy with which different gene targets represent community structure are not known. As a result, there is no consensus on which region provides the most suitable representation of diversity for eukaryotes. This study determined the accuracy with which commonly used 18S rRNA gene primer sets represent community structure and identified particular biases related to PCR amplification and Illumina MiSeq sequencing in order to more accurately study eukaryotic microbial communities.
INTRODUCTION
The use of high-throughput sequencing technologies (1, 2) has transformed the field of microbial ecology by contributing to a significant body of work that has changed our understanding of microbially diverse populations in a range of ecosystems. This is particularly true for investigations of bacterial and archaeal communities that target the 16S rRNA gene (1, 3–5). However, amplicon sequencing approaches for eukaryotes have lagged behind, due in part to the large variation in copy numbers of target genes among species (1 to >25,000 for the 18S rRNA gene) (6) and multiple hypervariable regions that are typically longer than early DNA sequencing platforms could sequence (7). The use of amplicon sequencing is of particular interest with respect to eukaryotic microalgae or phytoplankton due to their role in natural and engineered ecosystems (e.g., contribution to global carbon fixation [8], eutrophication of waterways [9, 10], treatment of nutrients and heavy metals in wastewater [11], and the production of biofuels [12], among others).
In engineered systems, microalgal technologies are uniquely positioned to provide solutions for both wastewater and energy industries by recovering nutrients (i.e., nitrogen [N] and phosphorus [P] via assimilation) and generating carbon-rich algal feedstock for downstream processing. Indeed, there is a growing consensus in the algal biofuel industry that wastewater should be leveraged to make algal biofuels environmentally and economically viable (13–15). Elucidating the relationship between system function (i.e., nutrient and carbon assimilation), operating parameters, and community composition requires a comprehensive examination of microalgal community structure (i.e., membership and relative abundance) and dynamics, the molecular tools for which are currently underdeveloped.
Although the wastewater field has used 16S rRNA amplicon sequencing to evaluate a wide range of bacterial communities (e.g., references 16 to 19), the use of high-throughput sequencing technologies with eukaryotic microalgae is virtually nonexistent, and the field relies heavily on microscopy for species identification. Sequencing offers the rapid detection of algal species without many of the problems associated with microscopy: (i) identification is not limited to those organisms with well-identified morphological markers; (ii) fewer personnel and less time are required (20); and (iii) hundreds of samples can be processed simultaneously by leveraging massively parallel approaches afforded by high-throughput sequencing. Furthermore, examining the community using amplicon sequencing allows us to take an in-depth look at the community structure of the organisms present. When coupled with other techniques, such as transcriptional analysis of a particular functional gene and/or statistical approaches to correlate reactor performance with the algal community, high-throughput sequencing may allow us to relate community structure to community function (21).
Despite recent studies that have developed broad eukaryotic primers (22, 23) using the small subunit (SSU) 18S rRNA gene, there is no widely accepted target gene used to sequence microalgae. Previous studies have targeted various regions of the rrn operon (e.g., 5.8S plus internal transcribed spacer 2 [ITS-2] [24, 25], 18S [8, 26–29], and 23S [30] regions), mitochondrial genes (e.g., cytochrome c oxidase 1 [COI] [31]), and chloroplast genes (e.g., the rbcL gene which encodes the large subunit for ribulose-1,5-bisphosphate carboxylase/oxygenase [RuBisCO] [32–34] and the 16S rRNA gene [35]), but these studies have been predominately limited to marine phytoplankton (e.g., references 29, 36, 37) and occasionally focused on freshwater phytoplankton (e.g., reference 38). The 18S rRNA gene is commonly amplified and offers the advantage of numerous alternating hypervariable (V1 to V9) and conserved regions. Within the 18S rRNA gene, multiple studies have used different variable regions for amplification, including the V1-V2 (39), V3 (40), V4 (41–43), and V9 (37, 41, 42) regions, with the V4 and V9 regions often being used together (e.g., references 41 and 42). A number of recent studies have compared variable regions along the entire 18S rRNA gene for all eukaryotes (22, 23) and eukaryotic plankton (44) and highlighted conserved regions that may be best suited for amplifying hypervariable regions. These studies identified primer combinations using in silico sequence database coverage and taxonomic resolution and confirmed their feasibility with environmental surveys. However, many of the regions identified by these studies are too long (>500 nucleotides [nt]) to allow for overlaps between the forward and reverse reads using the Illumina MiSeq platform (250- to 300-nt single read length, resulting in ∼450- to 500-nt-long combined reads with 50- to 150-bp overlap).
Additionally, a critical challenge that must be addressed is the quantification of errors associated with gene-based amplification (i.e., PCR bias) of these primer sets. There are several well-documented problems associated with the amplicon sequencing approach: (i) sequencing errors and chimeras may be formed during DNA amplification (45); (ii) primer coverage may not capture the desired microbially diverse populations (46); (iii) differential amplification efficiencies among the target gene may skew operational taxonomic unit (OTU) relative abundance (47, 48); and (iv) gene copy number variation may affect interpretations based on OTU relative abundance (49–51). The first issue has been the target of much research, particularly with respect to 16S rRNA gene sequencing (e.g., references 49 to 51), while previous research examining the coverage of 18S rRNA hypervariable regions (e.g., references 22, 23, and 44) provides insight into the second. Although PCR bias has been studied using 16S rRNA genes (e.g., references 52 and 53), the effect that it has on interpreting community structure has not been robustly addressed for the 18S rRNA gene.
This study seeks to address the effect of primer selection and resultant PCR/sequencing bias on the evaluation of eukaryotic microalgae (i.e., microalgae or phytoplankton) using 18S rRNA gene sequencing. In addition to identifying bias, we offer a redesigned primer set that solves problems associated with commonly used primers and more accurately represents microalgal communities in terms of coverage and relative abundance. Specifically, PCR/sequencing bias was examined by the following: (i) sequencing seven microalgal mock communities with different relative abundance constructs using 18S rRNA primers targeting the V4 and V8-V9 hypervariable regions, (ii) examining the variations in the detected community structure compared to the theoretical community structure by each primer set, (iii) quantifying the error in OTU mean relative abundance and its effect on alpha- and beta-diversity measurements, (iv) correlating the error in mean relative abundance to specific biases related to the PCR amplification process, and (v) testing improved primers on environmental samples for validation. In particular, we find that a commonly used V4 primer has critical shortcomings when applied to our mock community due to nucleotide mismatches on the 3′ end. The redesigned primer more accurately captures community structure and represents freshwater species with high accuracy, while the V8-V9 region offers good representation of all freshwater and marine species tested in this study.
MATERIALS AND METHODS
Primer design and evaluation.
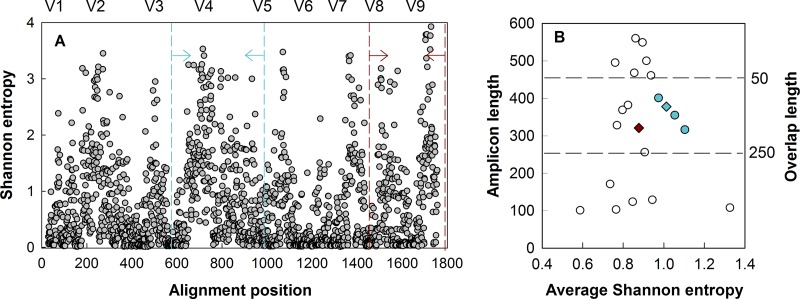
Primers were chosen by first examining the 18S rRNA gene through in silico testing. Database sequences for all eukaryotes were obtained from the SILVA database v119 (www.arb-silva.de, curated by mothur [54]), and trimmed to the S. cerevisiae reference sequence (accession number Z75578.1) using pcr.seqs in mothur v.1.34.0 (54). Shannon entropy was calculated according to the alignment position for all sequences (55) using the method of Shannon and the equation for each of the nucleotides present at a given location, where p(xj) was the frequency of the nucleotide xj at that alignment position (j). Following Shannon entropy calculations, conserved regions with alignment positions containing less than 0.2 entropy were identified (Fig. 1A).
FIG 1.
(A) Shannon entropy along the 18S rRNA gene alignment numbered according to corresponding positions in the 18S rRNA gene of Saccharomyces cerevisiae. (B) Average Shannon entropy per variable region compared to amplicon length with various primer combinations. Dashed lines in panel B indicate the overlap achievable with Illumina MiSeq v2 chemistry; diamonds indicate the primer sets selected for this study from the V4 (blue; alternative V4 primer sets are indicated by colored circles) and V8-V9 (red) hypervariable regions; the location of the selected set along the 18S rRNA gene is indicated by the dashed lines in panel A. The V4 region has the highest entropy within the overlap capabilities of the v2 chemistry, followed by the V8-V9 region. Although multiple primer sets are possible using the V4 region, we selected a set that has been used in previous studies. The V6 region is not highly variable and often is not included in discussion of the 18S hypervariable region.
Conserved regions were targeted as possible primer locations (22, 23, 41), and combinations of conserved/variable regions (i.e., possible amplicons) were plotted against Shannon entropy (Fig. 1B) in order to determine areas that had the highest entropy and amplicon length suitable for sequencing on the Illumina MiSeq platform. Conserved regions were then checked against general eukaryotic primers from the literature (Table 1) and examined for their suitability in amplifying the 18S rRNA gene. Priority was given to regions with high entropy and lengths compatible with current Illumina MiSeq sequencing (250 to 300 bp) to allow for maximal read overlap, and a primer set was selected from each V4 and V8-V9 region using established primers from the literature (references 41, 7, and 23, respectively) that have been used for numerous studies (37, 44, 56, 57). Although the V9 region (average 130 bp) (7) has typically been sequenced alone (7, 37), the V8 region was included to leverage the longer read capabilities that were not achievable with early high-throughput sequencing approaches (7). Selected primers were evaluated for general eukaryote- and alga-specific coverage using the SILVA TestPrime tool (58) (see Fig. S2 in the supplemental material).
TABLE 1.
Primers evaluated for 18S rRNA-based amplicon sequencing on the Illumina MiSeq platform
| Primer identification | S. cerevisiae position | Target region | Sequence | Reference or source |
|---|---|---|---|---|
| 550r | 550 | V4 | GGRCMAGBCTGGTGCCAG | 22 |
| 563f | 563 | V4 | GCCAGCAVCYGCGGTAAY | 22 |
| 574f | 574 | V4 | CGGTAAYTCCAGCTCYAV | 22 |
| Reuk454FWD1 | 565 | V4 | CCAGCASCYGCGGTAATTCC | 41a,b |
| ReukREV3 | 981 | V4 | ACTTTCGTTCTTGATYRA | 41a |
| V4r | 981 | V4 | ACTTTCGTTCTTGAT | This studyb |
| 1132r | 1150 | V4-V5 | CCGTCAATTHCTTYAART | 22 |
| V8f | 1422 | V8 | ATAACAGGTCTGTGATGCCCT | This studyb |
| 1422f | 1422 | V8 | ATAACAGGTCTGTGATGC | 23 |
| 1424f | 1424 | V8 | AACAGGTCHGWRATGCCC | 22 |
| 1510r | 1797 | V9 | CCTTCYGCAGGTTCACCTAC | 7b |
Primers initially selected for evaluation.
Primers selected for final sequencing run and recommended by this study.
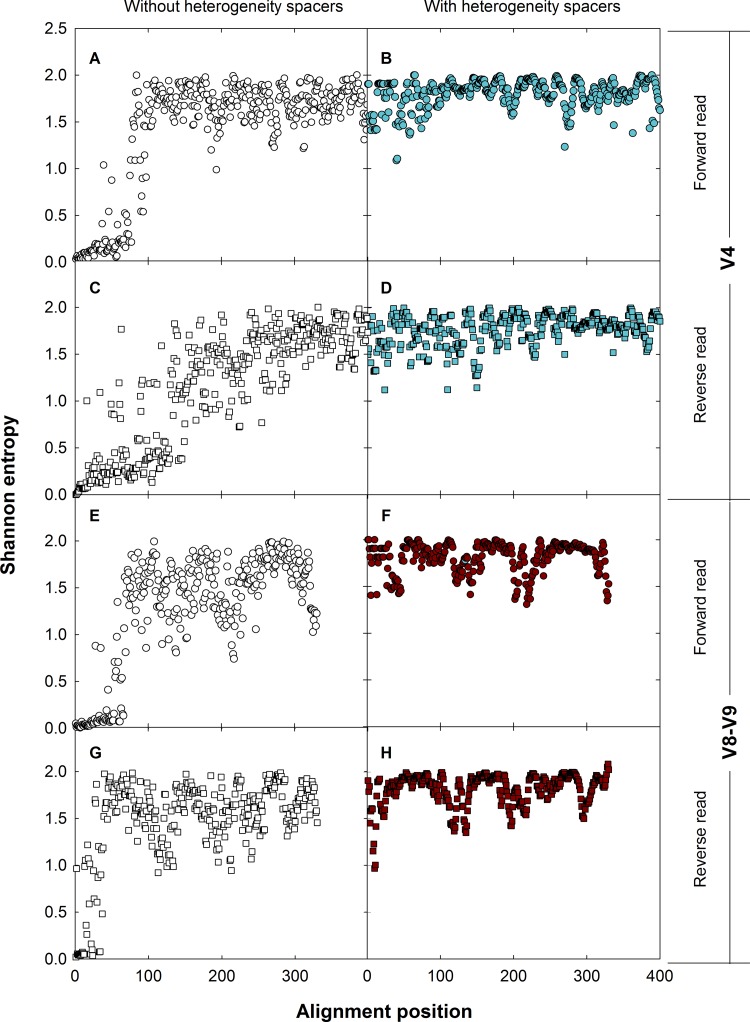
Full-length primers containing the adapters for Illumina MiSeq sequencing were constructed according to the dual-index method of Kozich et al. (59). Briefly, each forward and reverse primer consists of a 24- to 29-nucleotide-long Illumina MiSeq adapter to attach the DNA sequence to the MiSeq flow cell. The adapter is followed by an 8-nucleotide indexing sequence, a pad/linker sequence of 12 nucleotides to increase the overall melting temperature, and the 18S rRNA gene-specific primer. The presence of an indexing sequence on both the forward and reverse primers allows the multiplexed sequencing of a large number of samples with relatively few primers compared to traditional single-indexing methods that contain only one index on the forward or reverse primer (2). Based on the initial sequencing results (see Fig. S3 and S4 in the supplemental material), heterogeneity spacers, ranging from 0 to 7 nucleotides, were inserted between the indexing sequence and pad/linker region to phase the sequencing of conserved regions between samples and maximize “sequencing entropy” per cycle, similar to the method of Fadrosh et al. (60). This phasing approach allows better cluster delineation during the Illumina sequencing process, which may be particularly critical for low-diversity samples (61). Figure 2 shows the entropy per alignment position of the V4 and V8-V9 target regions with (Fig. 2B, D, F, and H) and without (Fig. 2A, C, E, and G) the heterogeneity spacers.
FIG 2.
Shannon entropy per alignment position without (open symbols) and with (filled symbols) heterogeneity spacers for V4 and V8-V9 as estimated using algal sequences extracted from the Silva 119 database: blue, V4; red, V8-V9; A, B, E, and F, forward read; C, D, G, and H, reverse read. Nucleotide diversity (as represented by Shannon entropy) is close to zero for the conserved primer regions and the reverse read of the V4 primer set (C) exhibits scattered entropy across the alignment. The addition of nucleotide spacers increases total entropy across the alignment for all primers.
DNA collection and extraction.
Axenic cultures for mock communities were obtained from the Culture Collection of Algae and Protozoa (CCAP) (Oban, United Kingdom) (see Table S1 in the supplemental material), the University of Texas Culture Collection of Algae (UTEX) (Austin, TX) (see Table S1 in the supplemental material), and the National Center for Marine Algae and Microbiota (NCMA) (see Table S1). Environmental samples for experimental validation were collected from freshwater, coastal, and wastewater sources from locations across the United States (see Table S2). DNA was extracted for all samples using a FastDNA SPIN extraction kit for soil (MP Biomedicals, Santa Ana, CA) and stored at −20°C until further processing.
Mock community construction.
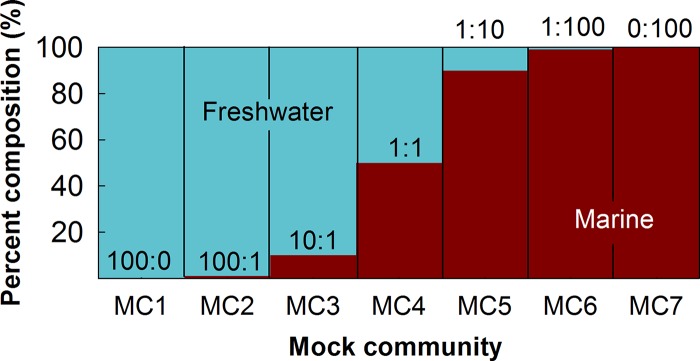
Mock communities of variable mean relative abundance were constructed from 12 algal species across 5 major divisions of eukaryotic microalgae of interest to the wastewater and biofuel field (Thalassiosira pseudonana, Chlorella vulgaris, Scenedesmus obliquus, Trebouxia sp., Cryptomonas pyrenoidifera, Rhodomonas sp., Heterocapsa niei, Symbiodinium microadriaticum, Prymnesium parvum, Isochrysis galbana, Ochromonas sp., and Nannochloropsis oculata) (for full details, see Table S1 in the supplemental material). Full-length 18S rRNA gene sequences were amplified via PCR with a Kapa HiFi HotStart PCR kit (Kapa Biosystems, Wilmington, MA) consisting of 1× Kapa HiFi buffer, 0.3 mM deoxynucleoside triphosphate (dNTP) mix, 0.3 μM forward/reverse primer, and 1 U HiFi HotStart DNA polymerase, with the addition of 10 ng template DNA per 50-μl reaction mixture with universal eukaryotic primers (62) EukA (5′-AACCTGGTTGATCCTGCCAGT-3′) and EukB (5′-TGATCCTTCTGCAGG-TTCACCTAC-3′) using standard desalted primers (Integrated DNA Technologies, Coralville, IA). PCR thermocycling conditions were as follows: 95°C for 10 min, 25 cycles of 95°C for 1 min, 65°C for 1.5 min, and 72°C for 2 min, with a final extension at 72°C for 10 min. The resultant PCR product was processed using gel electrophoresis, band extraction, and purification with a QIAquick gel purification kit (Qiagen, Valencia, CA) before being cloned via a TOPO TA clone kit (Invitrogen, Carlsbad, CA) with a pCR 4-TOPO TA vector, according to the manufacturer's protocols. The cloned PCR products were sequenced using Sanger sequencing and cloned plasmid primers M13For-20, M13Rev-21 (Invitrogen), and 563f (22). Individual Sanger reads were merged to obtain the full-length sequence, and species identification was confirmed using BLAST (63). Full-length 18S rRNA gene sequences are accessible from the National Center for Biotechnology Information (NCBI) GenBank database (accession numbers KU900218 to KU900229). The 12 species were categorized as either “freshwater” or “marine” based on their isolation source and mixed in equimolar amounts to create two groups of 6 species each. Species included in the marine mock community contain microalgae found in both open ocean and coastal environments. Seven mock communities (MC1-7) were created by combining the plasmid-cloned 18S rRNA gene sequences from each species in different freshwater/marine ratios (Fig. 3). The use of plasmid-cloned 18S rRNA gene sequences allowed for the addition of equal amounts of template to avoid variable gene copy numbers in the genomic DNA extracts from each organism.
FIG 3.
Composition of the seven mock communities, ranging from freshwater (MC1) to marine (MC7) only. Ratios show the theoretical freshwater/marine compositions of each community based on the number of 18S rRNA copies present.
DNA amplification and sequencing.
PCR was performed on all mock community and environmental samples in triplicate using high-performance liquid chromatography (HPLC)-purified dual-index barcoded primers (Eurofins MWG Operon, Huntsville, AL) (for the sequences of primer and indexing barcodes, see Tables S4 and S5 in the supplemental material). PCR was performed using the KAPA HiFi HotStart PCR kit (same concentrations as previously listed) using previously suggested thermocycling conditions for each hypervariable region. Specifically, for the V4 region (amended from Stoeck et al. [41]), the conditions were 95°C for 5 min, 10 cycles of 94°C for 30 s, 57°C for 45 s, and 72°C for 1 min, 15 cycles of 94°C for 30 s, 47°C for 45 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. For the V8-V9 region, the conditions were 95°C for 3 min, followed by 25 cycles of 98°C for 20 s, 65°C for 15 s, and 72°C for 15 s, with a final extension at 72°C for 10 min. A negative PCR control with no template DNA was included for each primer set. Gel electrophoresis was performed on all amplicons to confirm the amplicon size and quality before extraction and purification with a QIAquick gel purification kit (Qiagen). The DNA concentration of each sample amplicon library was checked with Qubit 2.0 (Invitrogen) in triplicate, followed by pooling of individual amplicon libraries in equimolar proportions.
Initial sequencing runs were performed by the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign (UIUC) and the Centre for Genomic Research at the University of Liverpool using Illumina MiSeq (Illumina, San Diego, CA) sequencing with v3 chemistry and 2 × 300 paired-end reads (sequencing runs 1 and 2, respectively). A subsequent run was performed by the Roy J. Carver Biotechnology Center at UIUC using the Illumina MiSeq with v2 chemistry and 2 × 250 paired-end reads (run 3). The read2 primer, consisting of a pad/linker sequence plus a V4 reverse primer, was created using HPLC-purified locked nucleic acids (LNA) (Exiqon A/S, Copenhagen, Denmark) in order to increase its melting temperature above that of the 65°C MiSeq cycling temperature to ensure nucleotide incorporation during sequencing. All other sequencing primers had melting temperatures above 65°C and were HPLC-purified oligonucleotides (Eurofins MWG Operon).
Data analysis.
Raw sequences were demultiplexed using bcl2fastq v1.8.4 Conversion Software (Illumina) before processing using Casava 1.8 (Illumina) and quality filtering using Sickle (64) to remove all bases with a phred score of less than 20 and to implement a minimum read overlap by specifying sequence read length. Sequences were then processed using mothur v1.34.0, following the method of Kozich et al. (59) (MiSeq SOP at http://www.mothur.org/wiki/MiSeq_SOP) and the default setting for all processing commands. After contig formation, reads with ambiguous base calls were removed, and sequences were trimmed to <400 and 350 bp using screen.seqs for the V4 and V8-V9 amplicons, respectively. Reads were aligned with the SILVA v119 NR alignment (provided by mothur), alignment was trimmed using vertical = T and trump =. options, and chimeras were detected in the trimmed alignment using UChime (65) and subsequently removed. Singletons were removed, and the remaining reads were used for all further analyses, including OTU clustering at various sequence similarity cutoffs (see Results and Discussion). A sequencing error was determined as the average percent difference between each mock community sequencing read and its reference Sanger sequence using seq.error. The consensus taxonomy of OTUs was performed using the Silva v119 taxonomy information provided by mothur. Alpha-diversity (observed OTUs, Chao1 index, inverse Simpson index, and nonparametric Shannon index) and beta-diversity (Jaccard and Bray-Curtis distances) metrics for the mock communities were calculated using the summary.single and summary.shared commands with 1,000 iterative subsampling efforts to the sample with the largest number of sequences that still allowed for replicate samples from each group (n = 5,489). Alpha-diversity metrics for the environmental samples were also calculated using summary.single and 1,000 iterative subsampling efforts to the largest number of sequences that still allowed for replicate samples from each group (n = 9,884). In order to directly compare the V4 and V8-V9 samples, the samples were binned into phylotypes using the phylogeny command, and beta-diversity metrics were calculated using dist.shared on a combined shared file. Scripts for processing these data in mothur have been uploaded to figshare and are accessible under https://dx.doi.org/10.6084/m9.figshare.3405577.v1.
Accession number(s).
Full-length 18S rRNA gene sequences for the individual mock community members are accessible from the National Center for Biotechnology Information (NCBI) GenBank database (accession numbers KU900218 to KU900229). Sequencing data used for analysis in this study are available through the NCBI Sequence Read Archive (SRA) (accession number SRP071862), and corresponding sample descriptions are accessible through BioProject PRJNA314977.
RESULTS AND DISCUSSION
Failed sequencing runs.
Despite the fact that the V4 primer set selected has been adapted to both the Roche 454 (41, 44) and the Illumina MiSeq (56) systems, an initial sequencing run (Illumina MiSeq, 2 × 300 paired-end reads) using the V4 primer set, which primes the reverse read and the indexing barcode associated with it for sequence identification, was unsuccessful due to a failure of the read2 and index1 primers (consisting of pad/linker plus V4 reverse primer). The read2/index primer was stripped off during the MiSeq's cycle chemistry because of a lower melting temperature (59.7 to 62.5°C) than the temperature at which nucleotides are incorporated during Illumina MiSeq sequencing (65°C). This resulted in no sequencing of the reverse strand or the reverse index (see Fig. S3 in the supplemental material). Although forward reads were obtained, they could not be assigned to samples because of the dual-indexing approach used.
Runs 2 and 3 utilized read2/index primers incorporating LNA to increase the primer-melting temperature to 70°C. The second sequencing run experienced two additional problems with the V4 primer set: low-quality reads resulting from poor cluster delineation caused by low-diversity environmental samples; and significant loss of microalgal coverage due to nucleotide mismatches on the 3′ end of the reverse primer. Microalgal organisms, particularly those present in wastewater treatment, are under highly selective pressures and, consequently, are often dominated by a few select organisms (66). Environmental samples selected for sequencing had highly similar nucleotide patterns, especially during the first ∼50 bp (see Fig. S4B in the supplemental material). Low diversity of nucleotides during the first 11 cycles is known to cause quality issues in the MiSeq because it uses these cycles to identify clusters and perform matrix calculations (61). The addition of PhiX to a sequencing run provides a control and generally improves nucleotide diversity; however, the reverse read during sequencing run 2 still showed poor quality despite the addition of PhiX (see Fig. S4A). A number of studies (e.g., references 60, 67, and 68) have proposed the use of variable-length nucleotide spacers to improve overall entropy and read quality of the sequencing library. An approach similar to that of Fadrosh et al. (60), whereby nucleotide spacers of different lengths were added to the indexing barcode, was used for run 3 and significantly improved the read quality of this sequencing run.
Additionally, the V4 primer set substantially altered the abundance representation of mock community members. Some species such as Rhodomonas sp. were overrepresented by approximately 4 to 5 times, while others were underrepresented by as much as 2 orders of magnitude (see Fig. S6 in the supplemental material). Two of the 12 members, Prymnesium parvum and Isochrysis galbana, were not detected at all. These errors in mean relative abundance were directly attributable to sequence read length and quality caused by nucleotide mismatches with the V4 reverse primer on the 3′ end. Meaningfully, the V4 reverse primer contained a degeneracy in the third nucleotide position and was made to match both 5′-TTG and 5′-TTA template sequences. Interestingly, full-length reads (380 bp) were only obtained for sequences containing the TTG motif at the priming location. Sequences with TTA corresponded to shortened reads (∼260 bp) and the underrepresentation of community members. The two species that were not represented had a complete mismatch on the 3′ end (CTG) and the shortest reads (<100 bp). The exact relationship between primer mismatches and resultant short amplicons will require further investigation. While it is known that primer mismatches disproportionately affect amplification efficiency when located near the 3′ end (69, 70), these data suggest that primers should be designed without degeneracies near the 3′ end as well. Differences in annealing temperature and specificity between G/C and A/T nucleotides may have resulted in the higher amplification of the TTG priming location (melting temperature [Tm] = 46.9°C) over that of TTA (Tm = 44.2°C), given the previously recommended thermocycling conditions used for PCR amplification in this study (minimum annealing temperature of 47°C). This discrepancy, along with the lack of coverage (which has been previously noted, e.g., reference 43) of the haptophytes Prymnesium parvum and Isochrysis galbana, represents a critical shortcoming of the V4 primer set as used in the literature. Consequently, this study used a “modified” V4 reverse primer (Table 1, V4r) without degeneracies on the 3′ end for the subsequent sequencing run.
Effect of read overlap and clustering similarities on sequence accuracy.
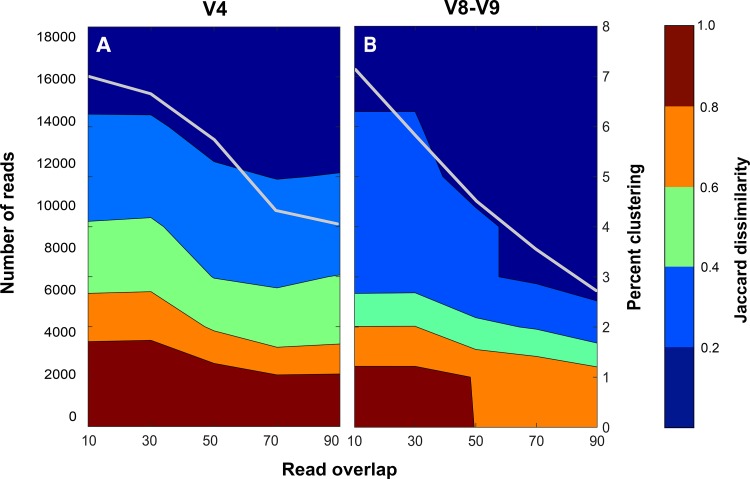
The lengths of forward and reverse reads (and, hence, read overlap) and clustering similarity affect the number of observed OTUs. It is common to use a similarity cutoff of 3 or 5% (71) to nominally express OTUs as taxonomic groupings at the species level, but it has been shown that a single threshold cannot be set to operationally define a clustering threshold and relate it to taxonomy (72). In order to determine the appropriate sequence similarity, clustering cutoffs to apply to this data set, read overlap (i.e., minimum read length specified during quality filtering), and clustering thresholds were varied at multiple cutoff values and compared to membership-based distance (i.e., Jaccard index) from the theoretical “even” mock community (Fig. 4). The goal of this analysis was to select the minimum read overlap and clustering threshold that accurately represented the mock community while retaining the most reads possible and reducing sequencing noise. As read overlap and similarity cutoff increased, the distance from the theoretical community decreased (i.e., increased in accuracy), but the number of reads included in the analyses also decreased. Using a cost-benefit relationship (the number of reads lost to the decrease in distance from theoretical), we determined the optimal cutoff values as the percentage of sequence similarity and read overlap at which the minimal number of reads was lost (i.e., cost) while decreasing the Jaccard distance from theoretical (i.e., benefit) to less than 0.2. For the V4 region, this was determined to be a 5% cutoff with a 70-bp overlap (225-bp read length). For the V8-V9 region, these criteria were satisfied at a 50-bp overlap at 5%. Although this similarity cutoff is higher than is often used (e.g., 3%), this analysis showed that a 5% threshold captured all mock community members with no loss of accuracy. By selecting a 5% cutoff, sequencing noise was masked, and spurious OTUs were reduced.
FIG 4.
Distance from the theoretical community (Jaccard dissimilarity) at various overlaps between forward and reverse reads and clustering of OTUs based on sequence similarity cutoffs for the V4 (A) and V8-V9 (B) regions using the “even” community (MC4). The left-hand y axis and gray line show the loss of reads as the minimum read overlap increases. Increasing the read overlap decreases the Jaccard dissimilarity but at the expense of loss of reads. The V4 and V8-V9 regions are able to achieve a Jaccard dissimilarity of <0.20 at 5% clustering (right-hand y axis) using 70- and 50-bp overlaps, respectively.
Examining the effect of read length and similarity cutoffs on community accuracy in this way also aided in identifying potential issues within the processed reads; local minimums (e.g., Fig. 4A: read overlap of 70 bp and sequence similarity of 3%) indicate regions where increasing the read overlap requirement actually increases the distance from theoretical, which might result from merging of paired-end reads that meet defined base quality metrics but are still sequencing errors. These points suggest that for those samples, read errors are making it through the quality filtering steps, and because the total number of sequences decreases with quality filtering, the effect of these sequencing errors is amplified. For example, increasing the read overlap from 70 to 80 bp for the even mock community (replicate 1 of 3) (see Fig. S7 in the supplemental material) resulted in 5 extraneous sequences and 2 additional OTUs being displayed (45 to 50 unique sequences, 13 to 15 OTUs for 70 and 80 bp, respectively).
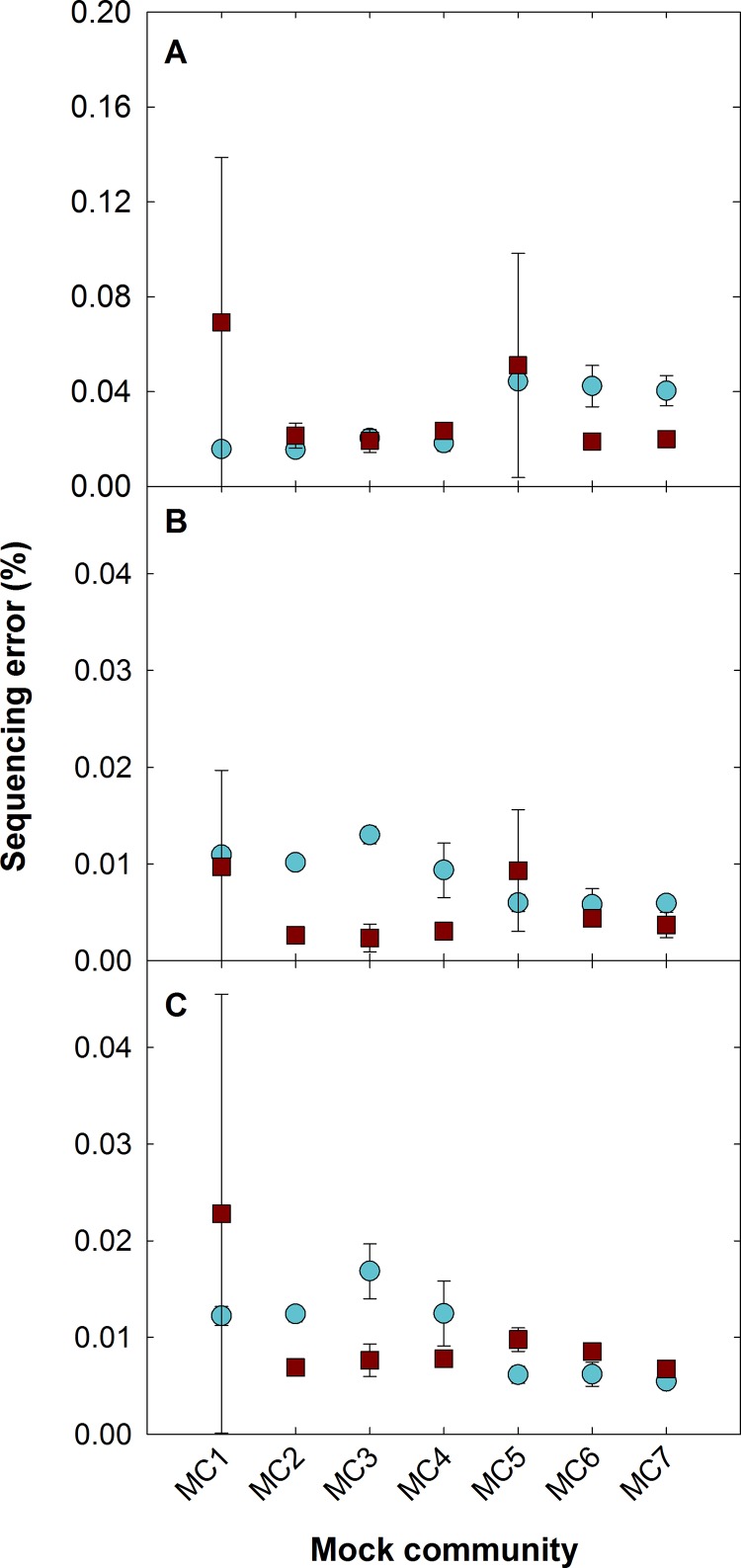
Sequencing error.
The V4 and V8-V9 primer sets show consistently low sequencing errors (i.e., the percent difference between mock community sequence reads and their known Sanger sequences) (Fig. 5, average 0.01% for processed sequences). Although there are limited data on acceptable error rates, Schloss et al. (73) evaluated multiple 16S rRNA primers and saw an average raw error of 0.61% across all regions and replicates before quality trimming and 0.56% after basic data processing such as removing ambiguous bases and instituting a minimum read length. This error was further reduced to 0.08% after application of a sliding window quality cutoff and sequence trimming. In comparison, all samples for the V4 and V8-V9 regions in this study show errors of <0.17% with only basic processing during the contig construction phase (i.e., no sequence trimming using quality scores). With quality trimming, all mock community samples fell below the 0.08% error seen by Schloss et al., with an average error of <0.024%.
FIG 5.
Effect of quality processing on sequencing errors for the V4 (blue) and V8-V9 (red) regions across the seven mock communities evaluated (MC1 to MC7). All reads were processed in mothur using no base quality cutoff (A), no base quality cutoff and singletons removed (B), and a base quality cutoff at a phred score of 20 and read overlaps of 70 bp (V4) and 50 bp (V8-V9) with singletons removed (C). Removing singletons has the greatest effect on reducing sequencing error.
Importantly, 77.1% of all mismatches in the V4 region were attributable to the two marine haptophytes Prymnesium parvum and Isochrysis galbana, even with the modified V4 reverse primer. Although overall error for the freshwater and marine communities was low, these members had error rates an order of magnitude higher than that for the overall marine community (2.82% compared to 0.04% for the overall community with basic processing). The haptophytes had a higher GC content for the V4 region than the rest of the marine community members (52% versus 44%), which has been shown to have a strong affect on PCR amplification (47, 74, 75). Consistent with this effect, all mismatches occurred in GC-rich regions and were predominantly substitution errors. Significantly, haptophytes have been shown to be routinely underrepresented when in the presence of other eukaryotic DNA (76, 77), and Marie et al. found that haptophytes present in marine samples had higher GC contents along the 18S rRNA gene than the other groups present (e.g., 49.2% for Haptophyta compared to 45.9% for Chlorophyta) (77).
Representation of mock communities.
In addition to the issues related to PCR priming locations and GC content of the template sequence (52, 75, 78), PCR amplification may be affected by (i) DNA template concentration (47), (ii) relative abundance, (iii) thermocycling conditions (79), (iv) primer choice, and (v) nonspecific binding that reduces PCR amplification (74). After the redesign of the V4 reverse primer, all mock community members shared exact priming sequences and item iv should not be a factor. It seems most likely that the GC content and nonspecific binding had the largest effect on mean relative abundance of community members.
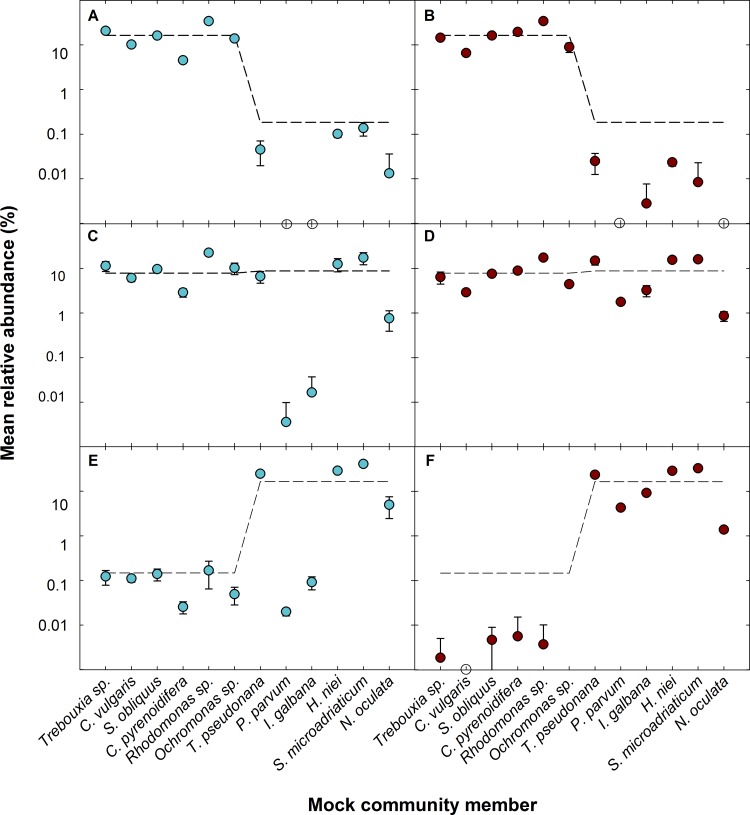
The mean relative abundance of 3 of the 7 mock communities (freshwater dominant, even, and marine dominant communities [MC2, MC4, and MC6, respectively]) is shown in Fig. 6 for the V4 and V8-V9 primer sets (for additional mock communities, see Fig. S8 in the supplemental material). Representation by the V8-V9 set outperforms that of the V4 set for all communities, with average mean relative abundances of 1.13-fold ± 0.32-fold and 1.61-fold ± 0.33-fold underrepresented per detected mock community member compared to the theoretical relative abundances for the V8-V9 and V4 amplicons, respectively. Although the V4 region captures freshwater species relatively well (0.28-fold ± 0.10-fold underrepresented from theoretical), it fails to adequately represent marine species (2.65-fold ± 0.33-fold underrepresented from theoretical), often underrepresenting the haptophyte members by as much as 3 orders of magnitude. Coupled with the higher sequencing error of the haptophytes, it is clear that the V4 region does not adequately capture the community structure of the marine mock community and will not accurately represent samples that contain these members. Unfortunately, haptophytes are of major importance to marine communities due to their contribution to open ocean biomass and production (76), and these studies most often leverage eukaryotic sequencing (e.g., references 41, 42, 57, and 80).
FIG 6.
Mean relative abundances of freshwater dominant (A and B), even (C and D), and marine dominant (E and F) communities corresponding to mock communities 2, 4, and 6 (MC2, MC4, and MC6), respectively, as represented by the V4 (A, C, and E; blue) and V8-V9 (B, D, and F; red) regions. The dashed lines indicate the mean relative abundance of the theoretical community. The V4 primer set consistently underrepresented the marine haptophyte members P. parvum and I. galbana. Although the V8-V9 primer set struggled to represent members when in low abundance, it more accurately represented the overall community structure.
Recent studies have highlighted the suitability of the V4 region for amplicon sequencing (22, 23), but these studies promote primer locations that include both the V4 and V5 regions (>500 bp long), which is longer than the Illumina MiSeq can currently sequence with paired-end reads (∼450 to 500 bp) with appropriate read overlap. Until longer reads are possible (and potentially even after), the V8-V9 region provides more accurate OTU relative abundances across all mock communities tested in this study. The V4 primer did capture both abundant and rare freshwater taxa, while the V8-V9 primer struggled with species (either freshwater or marine) that were at a low abundance. However, this is most likely an artifact of the amplification method used (i.e., concentrations of reagents, as well as the PCR cycle design), and there are multiple steps that can be taken to improve representation at low abundance. These include increasing primer and dNTP concentrations to allow for increased amplification of rare sequences, and the reduction of PCR cycles to minimize the overrepresentation of high-abundance sequences. Additionally, because most wastewater algal communities are under high selective pressures and are not very diverse, the V8-V9 primer set will reliably capture dominant organisms, while V4 may miss a dominant organism.
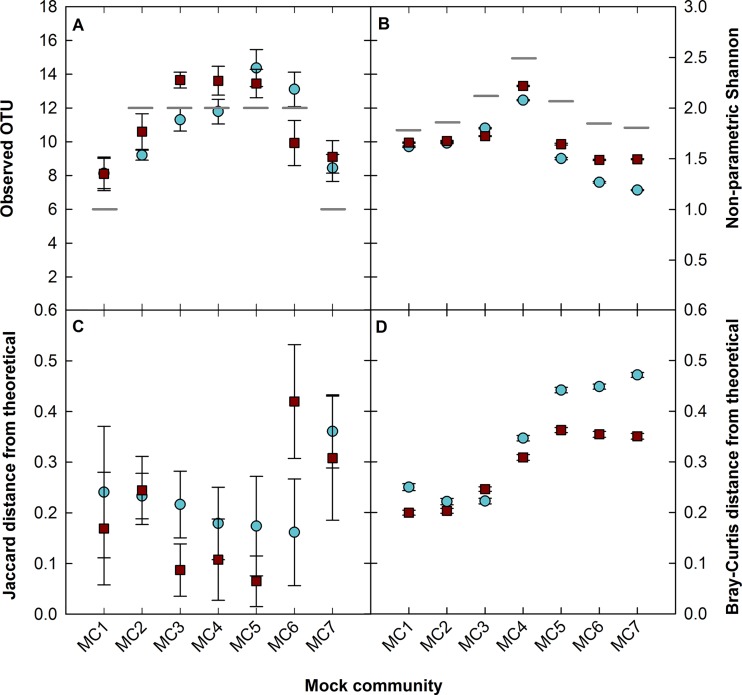
To further examine the performance of the V4 and V8-V9 regions, four alpha-diversity metrics were calculated: richness-based metrics (observed OTUs [SOBS], and the Chao1 [RCHAO] estimator, which estimate the unsampled richness); and structure-based metrics (nonparametric Shannon index [DNPSHANNON] and inverse Simpson index [DINVSIMPSON], which measure sample diversity assuming no underlying distribution and an even distribution, respectively). The V4 and V8-V9 regions show similar trends (P = 0.82 and 0.46, [Welch's t test]), with average SOBS and DNPSHANNON values (Fig. 7A and B) within 23% and 20% of the theoretical, respectively. Beta-diversity measurements, including the richness-based Jaccard (DJACCARD) and structure-based Bray-Curtis (DBRAYCURTIS) distances were also calculated. When the sequenced mock community distances from theoretical were compared, the V8-V9 region more accurately represented the theoretical community in 5 of 7 and 6 of 7 mock communities using Jaccard and Bray-Curtis metrics, respectively (Fig. 7C and D). Exceptions to this are DJACCARD for MC2 and MC6 due to some of the rare taxa not being detected. If low-abundance members were efficiently detected by incorporating the approaches discussed previously, it is likely that the DJACCARD from the theoretical community would also be smaller than that of the V4 region for these two communities.
FIG 7.
(A) Observed OTUs; (B) nonparametric Shannon index; (C) Jaccard distance from theoretical; (D) Bray-Curtis distance from theoretical. The V4 (blue) and V8-V9 (red) regions show similar alpha-diversity metrics in panels A and B (P = 0.82 and 0.46, respectively, Welch's t test; gray dashes indicate theoretical values), but the V8-V9 region more closely represents the theoretical community using beta-diversity metrics in panels C and D.
Application to environmental samples.
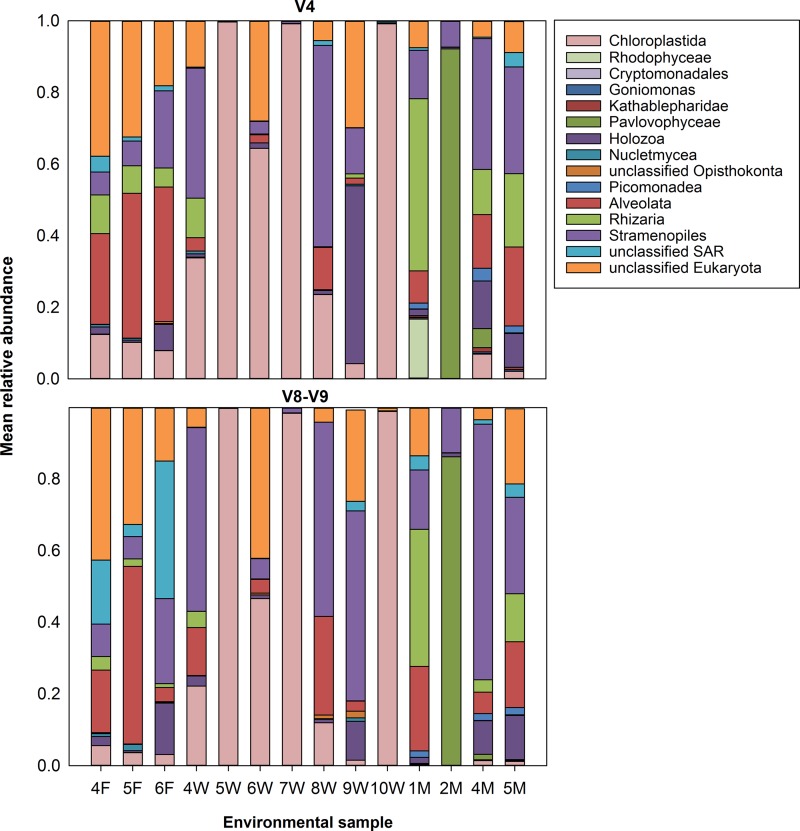
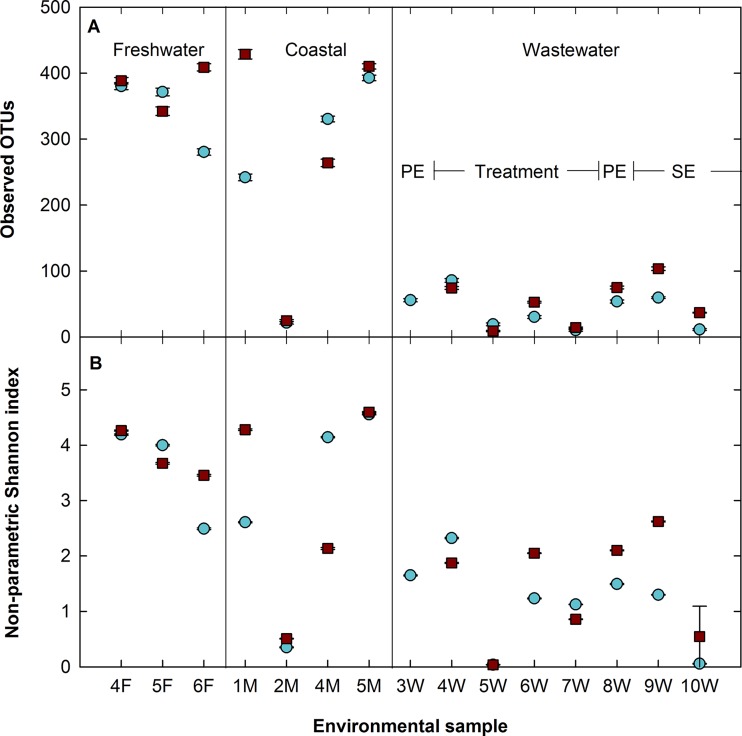
Community composition at taxonomic ranks of 3 and 6 in mothur (corresponding to class level[Fig. 8] and genus level [see Fig. S9 in the supplemental material], respectively) shows that microbial populations for each sample are similar as represented by either the V4 or V8-V9 region. Both the V4 and V8-V9 regions were able to differentiate among freshwater, coastal, and wastewater samples collected using Jaccard and Bray-Curtis metrics (P < 0.001, analysis of molecular variance [AMOVA]) (see Table S6 and Fig. S10 in the supplemental material). Furthermore, they were able to discriminate between communities within the wastewater treatment process, including primary clarification, secondary treatment, and secondary clarification (P < 0.005, AMOVA) (see Fig. S10). In general, the V8-V9 region had a greater number of observed OTUs and displayed higher levels of microbially diverse populations than the V4 region as estimated by using alpha-diversity metrics (DNPSHANNON) (Fig. 9).
FIG 8.
Community composition of the V4 (top) and V8-V9 (bottom) regions at a taxonomic rank of 3 in mothur, which corresponds to the class level. Samples containing F, W, and M in their sample name belong to freshwater, wastewater, and coastal marine samples, respectively. Data used to generate this figure are available in Table S7 in the supplemental material.
FIG 9.
Observed OTUs (A) and nonparametric Shannon index (B) of environmental samples by the V4 (blue) and V8-V9 (red) regions. Wastewater samples were taken from primary effluent (PE), treatment, or secondary effluent (SE) streams from wastewater treatment plants. Both hypervariable regions show significantly higher numbers of OTUs (P < 0.003, Welch's t test) in the freshwater and coastal samples than in wastewater and similar representations of metrics across all samples.
In order to examine the representation by each region of the coastal samples more closely, the Bray-Curtis distances between samples (e.g., 1M and 2M, 1M and 4M, etc.) were compared between the V4 and V8-V9 data sets. Welch's t tests (unpaired, two-tailed with unequal variance) showed that there were significant differences (P < 0.05) between the distances calculated among the samples from the V4 and V8-V9 regions, with each region giving different estimates of community dissimilarity between any two samples and the V4 data displaying higher dissimilarities than the V8-V9 data in 5 of the 6 pairwise comparisons. However, when taken collectively, the V4 and V8-V9 regions show similar trends overall, and a Mantel test between the V4 and V8-V9 Bray-Curtis distance matrices showed a high degree of similarity (0.90, P < 0.001). The V4 region had fewer OTUs for 3 of the 4 marine samples, but surprisingly, it represented organisms of the division Haptophyta as well as those of the V8-V9 region. Notably, the abundant genera found in environmental coastal samples all belonged to the class Pavlovophyceae, while the species present in the mock community belonged to the class Prymnesiophyceae. It is possible that the V4 region only struggles with representation of the latter, although additional mock community sequencing with members of the class Pavlovophyceae included would need to be performed.
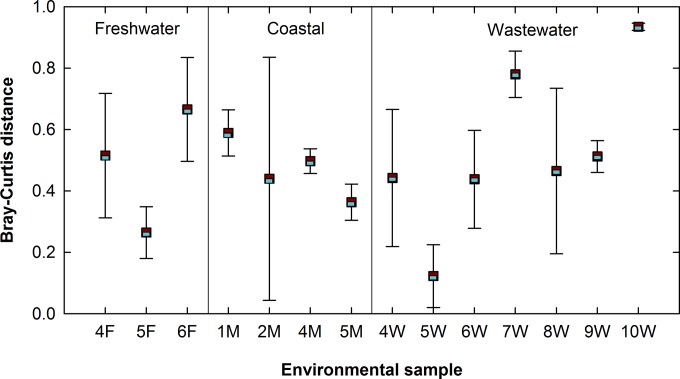
In order to directly compare differences in community representation by the V4 and V8-V9 regions, sequences were binned into phylotypes and beta-diversity metrics (Jaccard index and Bray-Curtis dissimilarity) were calculated. The V4 and V8-V9 regions show different community representations of the same sample (Fig. 10). The Bray-Curtis distance between the V4 community representation of a sample and its V8-V9 representation varies greatly, with some samples showing good agreement (5W, difference of 0.12) between both regions and others showing high dissimilarity (10W, difference of 0.94). Variations in the community representation by the V4 and V8-V9 regions do not correspond to sample type (i.e., freshwater, coastal, or wastewater), but rather, are sample and site specific. These results support the use of multiple primer sets as in previous studies (e.g., references 41 and 42) in order to provide a more complete picture of community representation.
FIG 10.
Bray-Curtis distance between the V4 and V8-V9 representations of the same sample community using a phylotype-based approach. Community representation by the V4 and V8-V9 regions varies in agreement on a sample-by-sample basis, rather than across sample types (i.e., freshwater, coastal, or wastewater).
Despite differences in community representation by each hypervariable region (as measured through the Bray-Curtis distance), the three data sets cluster according to their sample types irrespective of the hypervariable region targeted (see Fig. S11 in the supplemental material). Furthermore, although samples can be differentiated according to sample type (e.g., freshwater or wastewater; P < 0.001, AMOVA), there is no significant difference between the V4 samples and V8-V9 samples within the same type of environmental sample (e.g., V4 freshwater and V8-V9 freshwater; P > 0.05, AMOVA).
In conclusion, in this study, we examined the effect of PCR/sequencing bias on the representation of seven mock communities by targeting the V4 and V8-V9 hypervariable regions of the 18S rRNA gene. By doing so, we discovered that a previously used primer set missed a major taxonomic group of interest to marine studies. This study highlights the need for mock communities to validate the representation of species by amplicon sequencing; in silico testing can help identify sequence coverage and nucleotide mismatches, but experimental validation with mock communities provides critical insight into the amplification, sequencing, and representation of target regions. Specifically, this study found that nucleotide degeneracies on the primer 3′ end affected read lengths and mean relative abundances of mock communities due to differential amplification of templates containing G or A in the degenerate position for V4 primers proposed in literature. Furthermore, the PCR/sequencing error is markedly higher for GC-rich members (2.82% compared to an average 0.04% for mock community 4). Importantly, the V4 region failed to reliably capture 2 of the 12 mock community members included in this study. The V8-V9 region more accurately represents mean relative abundance and alpha and beta diversity, with the greatest improvement in structure-based metrics such as the Bray-Curtis distance from theoretical.
Given the additional uncertainty of gene copy numbers across eukaryotic species, representation of even closely related organisms (such as phytoplankton) by amplicon sequencing has a high degree of ambiguity. Complementary approaches such as application of diversity metrics only to similarly sized microalgae (37) or sequencing cell-sorted populations (77) might greatly improve the accuracy of diversity metrics and mean relative abundance. In the case of Marie et al., the use of cell sorting allowed for the sequencing of abundant phytoplankton (e.g., Haptophyta) that were previously undetected due to the profusion of larger organisms (77). The effect of PCR bias, however, skews diversity estimates even when the gene copy number is kept the same. Based on these results, we recommend that studies that apply amplicon sequencing to environmental samples do the following:(i) use mock communities composed of target species to estimate community representation and PCR/sequencing error by the chosen primer set; (ii) limit degenerate locations in primer sequences and eliminate degeneracies in positions near the 3′ end; and (iii) estimate alpha- and beta-diversity metrics through structure-based methods that provide more reliable approximations of community diversity.
In the current study, redesign of the primer containing a degeneracy on the 3′ end increased the representation of the marine members and improved mock community representation overall. Ultimately, the V8-V9 region provided the highest accuracy of the selected mock community as measured through mean relative abundance and beta-diversity measurements (DJACCARD and DBRAYCURTIS). Given these data, we suggest that studies using 18S rRNA gene amplicon sequencing for microalgal communities (and marine studies in particular) target the V8-V9 hypervariable region when considering species included in this study. However, the V4 and V8-V9 regions showed similar overall representations of environmental samples and tradeoffs between hypervariable regions may warrant the use of multiple primer sets to better capture community diversity.
Supplementary Material
ACKNOWLEDGMENTS
This material is based upon work supported by the National Science Foundation (grant CBET-1351667) and the Engineering and Physical Sciences Research Council, United Kingdom (grant EP/K035886/1).
We acknowledge the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign (UIUC) and the Centre for Genomic Research at the University of Liverpool for performing high-throughput sequencing. We thank Andrew Henderson, Belinda Sturm, Daniel Johnson, Lance Schideman, Chih-Ting Kuo, Monica Higgins, Diego Rosso, Graham Mccarthy, Andrew Whelton, and Carolina Novy for generously donating environmental samples for this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01630-16.
REFERENCES
- 1.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103:12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teske A, Hinrichs K-U, Edgcomb V, de Vera Gomez A, Kysela D, Sylva SP, Sogin ML, Jannasch HW. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol 68:1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prokopowich C, Gregory T, Crease T. 2003. The correlation between rDNA copy number and genome size in eukaryotes. Genome 46:48−50. doi: 10.1139/g02-103. [DOI] [PubMed] [Google Scholar]
- 7.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viprey M, Guillou L, Ferréol M, Vaulot D. 2008. Wide genetic diversity of picoplanktonic green algae (Chloroplastida) in the Mediterranean Sea uncovered by a phylum-biased PCR approach. Environ Microbiol 10:1804–1822. doi: 10.1111/j.1462-2920.2008.01602.x. [DOI] [PubMed] [Google Scholar]
- 9.Conley DJ, Paerl HW, Howarth RW, Boesch DF, Seitzinger SP, Havens KE, Lancelot C, Likens GE. 2009. Ecology-controlling eutrophication: nitrogen and phosphorus. Science 323:1014–1015. doi: 10.1126/science.1167755. [DOI] [PubMed] [Google Scholar]
- 10.Paerl HW, Valdes LM, Joyner AR, Piehler MF, Lebo ME. 2004. Solving problems resulting from solutions: evolution of a dual nutrient management strategy for the eutrophying Neuse River Estuary, North Carolina. Environ Sci Technol 38:3068–3073. doi: 10.1021/es0352350. [DOI] [PubMed] [Google Scholar]
- 11.Craggs RJ, Davies-Colley RJ, Tanner CC, Sukias JP. 2003. Advanced pond system: performance with high rate ponds of different depths and areas. Water Sci Technol 48:259–267. [PubMed] [Google Scholar]
- 12.Chisti Y. 2007. Biodiesel from microalgae. Biotechnol Adv 25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Clarens AF, Resurreccion EP, White MA, Colosi LM. 2010. Environmental life cycle comparison of algae to other bioenergy feedstocks. Environ Sci Technol 44:1813–1819. doi: 10.1021/es902838n. [DOI] [PubMed] [Google Scholar]
- 14.Christenson L, Sims R. 2011. Production and harvesting of microalgae for wastewater treatment, biofuels, and bioproducts. Biotechnol Adv 29:686–702. doi: 10.1016/j.biotechadv.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Pittman JK, Dean AP, Osundeko O. 2011. The potential of sustainable algal biofuel production using wastewater resources. Bioresour Technol 102:17–25. doi: 10.1016/j.biortech.2010.06.035. [DOI] [PubMed] [Google Scholar]
- 16.Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops H-P, Wagner M. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382. doi: 10.1128/AEM.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer KH. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol 63:2884–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Röser A, Koops HP, Wagner M. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol 64:3042–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crocetti GR, Hugenholtz P, Bond PL, Schuler A, Keller J, Jenkins D, Blackall LL. 2000. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl Environ Microbiol 66:1175–1182. doi: 10.1128/AEM.66.3.1175-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eland LE, Davenport R, Mota CR. 2012. Evaluation of DNA extraction methods for freshwater eukaryotic microalgae. Water Res 46:5355–5364. doi: 10.1016/j.watres.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 21.de los Reyes FL. 2010. Challenges in determining causation in structure-function studies using molecular biological techniques. Water Res 44:4948–4957. doi: 10.1016/j.watres.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 22.Hugerth LW, Muller EEL, Hu YOO, Lebrun LA, Roume MH, Lundin D, Wilmes P, Andersson AF. 2014. Systematic design of 18S rRNA gene primers for determining eukaryotic diversity in microbial consortia. PLoS One 9:e95567. doi: 10.1371/journal.pone.0095567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadziavdic K, Lekang K, Lanzen A. 2014. Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS One 9:e87624. doi: 10.1371/journal.pone.0087624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundholm N, Moestrup O, Kotaki Y, Hoef-Emden K, Scholin C, Miller P. 2006. Inter-and intraspecific variation of the Pseudo-nitzschia delicatissima complex (Bacillariophyceae) illustrated by rRNA probes, morphological data and phylogenetic analyses. J Phycol 42:464–481. doi: 10.1111/j.1529-8817.2006.00211.x. [DOI] [Google Scholar]
- 25.Guo L, Sui Z, Zhang S, Ren Y, Liu Y. 2015. Comparison of potential diatom “barcode” genes (18S and ITS rDNA, COI, rbcL) and their effectiveness in discriminating and determining species taxonomy in Bacillariophyta. Int J Syst Evol Microbiol 65(Pt 4):1369–1380. doi: 10.1099/ijs.0.000076. [DOI] [PubMed] [Google Scholar]
- 26.Medlin LK, Kooistra WHCF, Gersonde R, Wellbrock U. 1996. Evolution of the diatoms (Bacillariophyta). II. Nuclear-encoded small-subunit rRNA sequence comparisons confirm a paraphyletic origin for the centric diatoms. Mol Biol Evol 13:67–75. [DOI] [PubMed] [Google Scholar]
- 27.Zhu F, Massana R, Not F, Marie D, Vaulot D. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Bazin P, Jouenne F, Deton-Cabanillas A-F, Pérez-Ruzafa Á, Véron B. 2013. Complex patterns in phytoplankton and microeukaryote diversity along the estuarine continuum. Hydrobiologia 726:155–178. [Google Scholar]
- 29.Di B, Pedrós-alió C, Massana R. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol 67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherwood AR, Presting GG. 2007. Universal primers amplify a 23S rDNA plastid marker in eukaryotic algae and cyanobacteria. J Phycol 43:605–608. doi: 10.1111/j.1529-8817.2007.00341.x. [DOI] [Google Scholar]
- 31.Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc Biol Sci 270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh S, Love NG. 2011. Application of rbcL based molecular diversity analysis to algae in wastewater treatment plants. Bioresour Technol 102:3619–3622. doi: 10.1016/j.biortech.2010.10.125. [DOI] [PubMed] [Google Scholar]
- 33.Paul JH, Alfreider A, Wawrik B. 2000. Micro- and macrodiversity in rbcL sequences in ambient phytoplankton populations from the southeastern Gulf of Mexico. Mar Ecol Prog Ser 198:9–18. doi: 10.3354/meps198009. [DOI] [Google Scholar]
- 34.Hepperle D, Krienitz L. 2001. Systematics and ecology of chlorophyte picoplankton in German inland waters along a nutrient gradient. Int Rev Hydrobiol 86:269–284. doi:. [DOI] [Google Scholar]
- 35.Fuller NJ, Campbell C, Allen DJ, Pitt FD, Zwirglmaier K, Le Gall F, Vaulot D, Scanlan DJ. 2006. Analysis of photosynthetic picoeukaryote diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquat Microb Ecol 43:79–93. doi: 10.3354/ame043079. [DOI] [Google Scholar]
- 36.Vaulot D, Eikrem W, Viprey M, Moreau H. 2008. The diversity of small eukaryotic phytoplankton (< or =3 microm) in marine ecosystems. FEMS Microbiol Rev 32:795–820. doi: 10.1111/j.1574-6976.2008.00121.x. [DOI] [PubMed] [Google Scholar]
- 37.de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, Carmichael M, Poulain J, Romac S, Colin S, Aury JM, Bittner L, Chaffron S, Dunthorn M, Engelen S, Flegontova O, Guidi L, Horák A, Jaillon O, Lima-Mendez G, Lukeš J, Malviya S, Morard R, Mulot M, Scalco E, Siano R, Vincent F, Zingone A, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Tara Oceans Coordinators, Acinas SG, Bork P, Bowler C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Not F, Ogata H, Pesant S, Raes J, Sieracki ME, Speich S, Stemmann L, Sunagawa S, Weissenbach J, Wincker P, Karsenti E. 2015. Eukaryotic plankton diversity in the sunlit ocean. Science 348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 38.Eiler A, Drakare S, Bertilsson S, Pernthaler J, Peura S, Rofner C, Simek K, Yang Y, Znachor P, Lindström ES. 2013. Unveiling distribution patterns of freshwater phytoplankton by a next generation sequencing based approach. PLoS One 8:e53516. doi: 10.1371/journal.pone.0053516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohrbeck I, Raupach MJ, Martínez Arbizu P, Knebelsberger T, Laakmann S. 2015. High-throughput sequencing—the key to rapid biodiversity assessment of marine metazoa? PLoS One 10:e0140342. doi: 10.1371/journal.pone.0140342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medinger R, Nolte V, Pandey RV, Jost S, Ottenwälder B, Schlötterer C, Boenigk J. 2010. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol 19(Suppl 1):S32–S40. doi: 10.1111/j.1365-294X.2009.04478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner H-W, Richards TA. 2010. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol 19(Suppl 1):S21–S31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 42.Pagenkopp Lohan KM, Fleischer RC, Carney KJ, Holzer KK, Ruiz GM. 2016. Amplicon-based pyrosequencing reveals high diversity of protistan parasites in ships' ballast water: implications for biogeography and infectious diseases. Microb Ecol 71:530−542. doi: 10.1007/s00248-015-0684-6. [DOI] [PubMed] [Google Scholar]
- 43.Balzano S, Abs E, Leterme SC. 2015. Protist diversity along a salinity gradient in a coastal lagoon. Aquat Microb Ecol 74:263–277. doi: 10.3354/ame01740. [DOI] [Google Scholar]
- 44.Tanabe AS, Nagai S, Hida K, Yasuike M, Fujiwara A, Nakamura Y, Takano Y, Katakura S. 2016. Comparative study of the validity of three regions of the 18S-rRNA gene for massively parallel sequencing-based monitoring of the planktonic eukaryote community. Mol Ecol Resour 16:402−414. doi: 10.1111/1755-0998.12459. [DOI] [PubMed] [Google Scholar]
- 45.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 46.Hong S, Bunge J, Leslin C, Jeon S, Epstein SS. 2009. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J 3:1365–1373. doi: 10.1038/ismej.2009.89. [DOI] [PubMed] [Google Scholar]
- 47.Polz MF, Cavanaugh CM. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64:3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki MT, Giovannoni SJ. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62:625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods 6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 50.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinto AJ, Raskin L. 2012. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7:e43093. doi: 10.1371/journal.pone.0043093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou J, Wu L, Deng Y, Zhi X, Jiang Y, Tu Q, Xie J, Van Nostrand JD, He Z, Yang Y. 2011. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 5:1303–1313. doi: 10.1038/ismej.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon CE. 1948. A mathematical theory of communication. Bell Syst Technol J 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 56.Carney LT, Reinsch SS, Lane PD, Solberg OD, Jansen LS, Williams KP, Trent JD, Lane TW. 2014. Microbiome analysis of a microalgal mass culture growing in municipal wastewater in a prototype OMEGA photobioreactor. Algal Res 4:52–61. doi: 10.1016/j.algal.2013.11.006. [DOI] [Google Scholar]
- 57.Logares R, Audic S, Santini S, Pernice MC, de Vargas C, Massana R. 2012. Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing. ISME J 6:1823–1833. doi: 10.1038/ismej.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. 2014. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Illumina. 2013. Low-diversity sequencing on the Illumina. Illumina, San Diego, CA. [Google Scholar]
- 62.Medlin L, Elwood H, Stickel S, Sogin M. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 63.Altschul S. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.Joshi N, Fass J. 2011. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33). https://github.com/najoshi/sickle.
- 65.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park JBK, Craggs RJ, Shilton AN. 2011. Recycling algae to improve species control and harvest efficiency from a high rate algal pond. Water Res 45:6637–6649. doi: 10.1016/j.watres.2011.09.042. [DOI] [PubMed] [Google Scholar]
- 67.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. 2013. Practical innovations for high-throughput amplicon sequencing. Nat Methods 10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 68.Wu L, Wen C, Qin Y, Yin H, Tu Q, Van Nostrand JD, Yuan T, Yuan M, Deng Y, Zhou J. 2015. Phasing amplicon sequencing on Illumina MiSeq for robust environmental microbial community analysis. BMC Microbiol 15:125. doi: 10.1186/s12866-015-0450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stadhouders R, Pas SD, Anber J, Voermans J, Mes THM, Schutten M. 2010. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J Mol Diagn 12:109–117. doi: 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bru D, Martin-Laurent F, Philippot L. 2008. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl Environ Microbiol 74:1660–1663. doi: 10.1128/AEM.02403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci 361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reysenbach AL, Giver LJ, Wickham GS, Pace NR. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol 58:3417−3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benita Y, Oosting RS, Lok MC, Wise MJ, Humphery-Smith I. 2003. Regionalized GC content of template DNA as a predictor of PCR success. Nucleic Acids Res 31:e99. doi: 10.1093/nar/gng101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moon-van der Staay SY, van der Staay GWM, Guillou L, Vaulot D, Claustre H, Medlin LK. 2000. Abundance and diversity of prymnesiophytes in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol Oceanogr 45:98–109. doi: 10.4319/lo.2000.45.1.0098. [DOI] [Google Scholar]
- 77.Marie D, Shi XL, Rigaut-Jalabert F, Vaulot D. 2010. Use of flow cytometric sorting to better assess the diversity of small photosynthetic eukaryotes in the English Channel. FEMS Microbiol Ecol 72:165–178. doi: 10.1111/j.1574-6941.2010.00842.x. [DOI] [PubMed] [Google Scholar]
- 78.Mamedov TG, Pienaar E, Whitney SE, TerMaat JR, Carvill G, Goliath R, Subramanian A, Viljoen HJ. 2008. A fundamental study of the PCR amplification of GC-rich DNA templates. Comput Biol Chem 32:452–457. doi: 10.1016/j.compbiolchem.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ishii K, Fukui M. 2001. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl Environ Microbiol 67:3753–3755. doi: 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duret MT, Pachiadaki MG, Stewart FJ, Sarode N, Christaki U, Monchy S, Srivastava A, Edgcomb VP. 2015. Size-fractionated diversity of eukaryotic microbial communities in the Eastern Tropical North Pacific oxygen minimum zone. FEMS Microbiol Ecol 91:fiv037. doi: 10.1093/femsec/fiv037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.