Fig. 4.
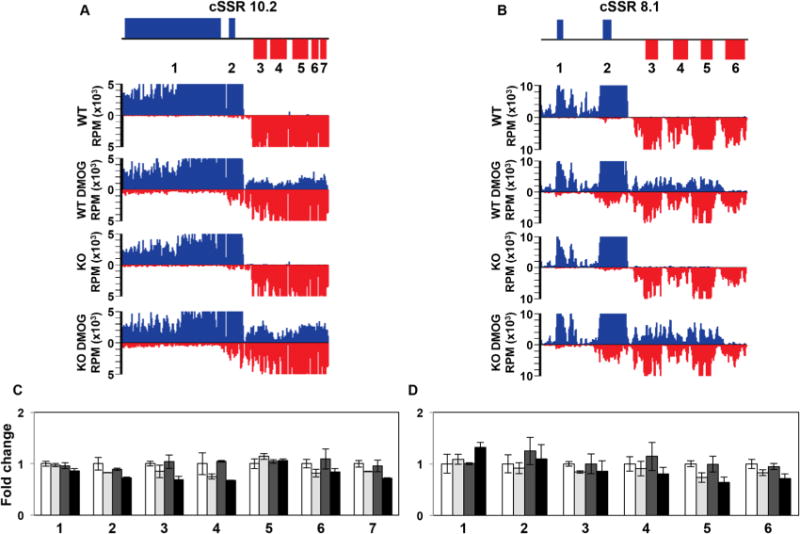
Read through transcription does not lead to transcriptional interference.
A and B. Strand-specific mRNA-seq reads for two representative cSSRs where loss of J leads to high degree of read through transcription into the adjacent gene cluster are shown. The genomic region shown is from 243 to 287 kb on chromosome 10 in (A) and from 384 to 406 kb on chromosome 8 in (B).
C and D. Plot of the mRNA-seq data for the genes indicated (numbered) in the ORF map for the corresponding regions above. The average RPKM of mRNA-seq replicate libraries was used to determine the fold changes, with WT set to one. Error bars indicate the standard deviation between mRNA-seq replicates. White bars: Wild type; grey bars: Wild type+DMOG; dark grey bars: H3.V KO; black bars: H3.V KO+DMOG. None of the genes shown are significantly differentially expressed relative to WT except for gene 5 (LmjF.08.0890) in D in the H3.V KO+DMOG condition, as determined by Cuffdiff (P-value of 0.03), but gene 5 is only downregulated by 1.6 fold, which does not meet our twofold cutoff.
