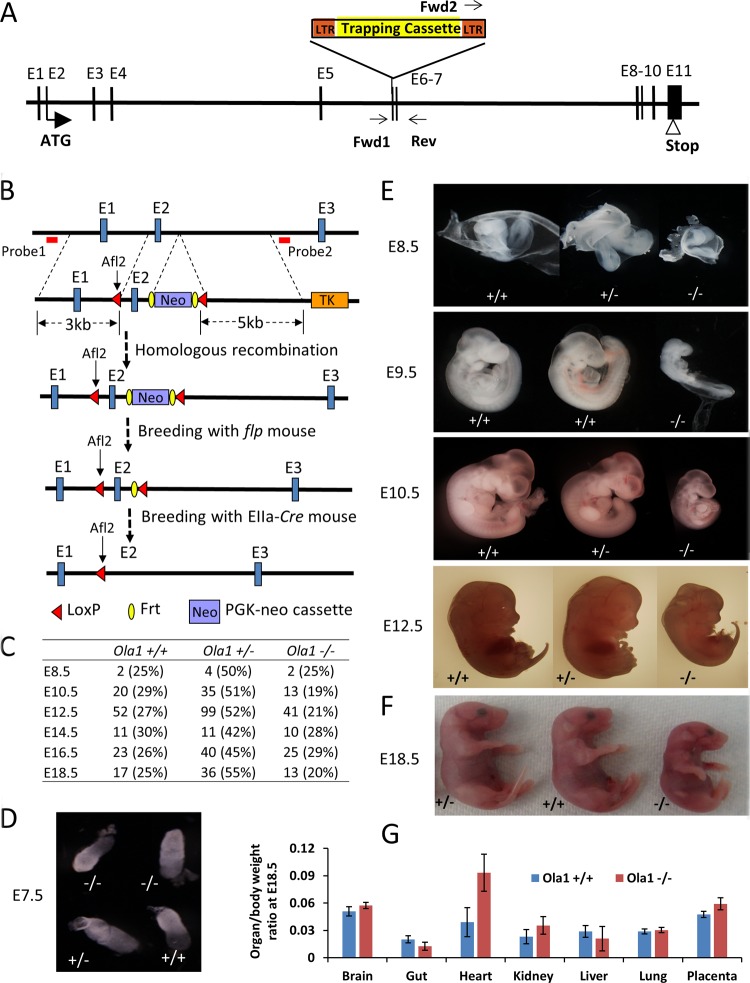
FIG 1.
Targeted disruption of Ola1 causes growth retardation and developmental delay. (A) Gene trapping at the Ola1 locus. A retroviral vector (Omni Bank VICTR48) was inserted at exon 6 (E6, or the 5th coding exon) of the Ola1 gene. This insertion was identified using PCR with primers Forward 1, Forward 2, and Reverse, as indicated. LTR, long terminal repeat. (B) Deletion of exon 2 (E2) using a Cre-loxP strategy. A targeting construct was designed to introduce a loxP site to intron 1 and a PGK-neo cassette flanked by FRT that was linked to the 2nd loxP (top). It also contained a 3-kb 5′ homology arm, a 5-kb 3′ arm, and the herpes simplex virus thymidine kinase (TK) gene, which was used as a negative selection marker. The positions of the 5′ and 3′ external probes that were used for Southern blot analysis are shown as red bars. The first loxP was fused with a rare-cutting restriction enzyme site (Alf2), and the presence of this site was identified using Southern blot analysis or PCR followed by Alf2 digestion. The FRT-PGK-neo-FRT cassette served as the positive selection marker. The correctly targeted ES cells, carrying the knock-in allele, were used to produce mice heterozygous for the allele (2nd panel). By breeding these mice with flpe transgenic mice, the neo gene was deleted, resulting in floxed mice (Ola1f/+) (3rd panel). Finally, by breeding Ola1f/f mice with EIIa-Cre mice, mice heterozygous for the Cre-recombined allele (Ola1f/−) were generated (bottom panel). (C) Genotypes of the progeny of Ola1 heterozygous intercrosses. Shown are the numbers of embryos observed, with the percentages shown in parentheses. (D to F) Morphological examination of Ola1+/+, Ola1+/−, and Ola1−/− embryos at E7.5 (D), from E8.5 to E12.5 (E), and at E18.5 (F). (G) Ratios of organ weight to body weight are shown for various internal organs and the placenta for E18.5 Ola1+/+ and Ola1−/− fetuses. The data are presented as the means ± SEMs (n = 8 to 16).