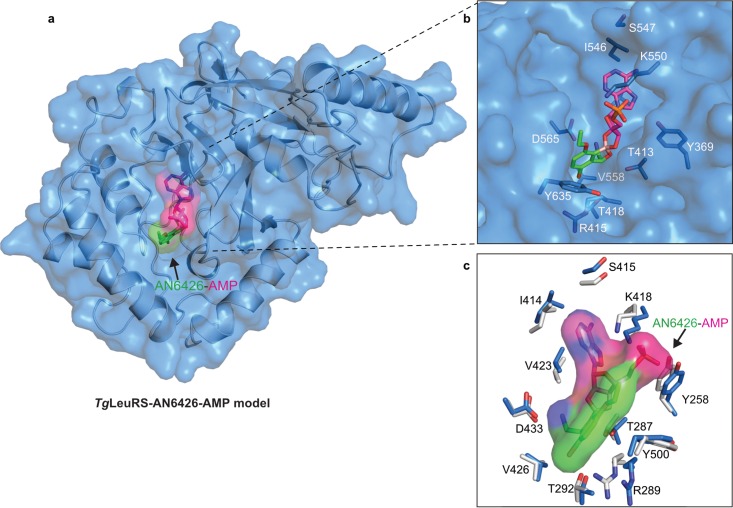
FIG 4.
TgLeuRS-AN6426-AMP model and conservation of residues. (a) Homology model of TgLeuRS calculated by structural alignment and subsequent model building using as a template the editing domain of the CmLeuRS complex with AN6426-AMP determined in this work (see Materials and Methods). The protein model is shown in a carton-and-surface representation (blue); the inhibition adduct is shown in a sticks-and-surface representation, with the AMP moiety being shown in pink and AN6426 being shown in green. (b) Magnification of the view of AN6426-AMP in the editing site of TgLeuRS, which shows no steric clashes with residues that are key to the interaction with AN6426-AMP (shown as blue sticks). (c) Conservation of residues between the editing sites of CmLeuRS and TgLeuRS. Key residues in the interaction with AN6426-AMP (shown in a sticks-and-surface representation) are shown as blue sticks for TgLeuRS and white sticks for TgLeuRS. For clarity, only residues of CmLeuRS are labeled. The figure is rotated 180 degrees with respect to the orientation in panel b.