Fig. 5. Nej1 stimulates gap-filling DNA synthesis by Pol4.
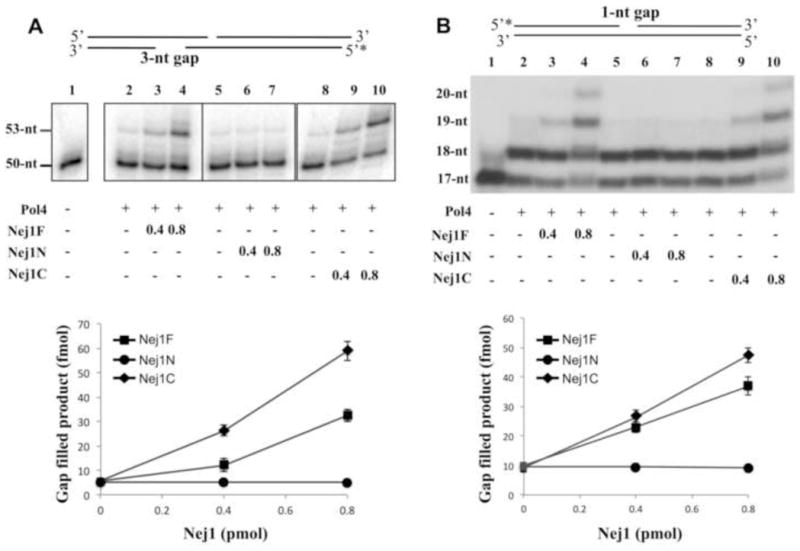
(A) One of two linear DNA duplexes with partially complementary ends that anneal to form a 3-nucleotide gap was end-labeled as indicated (0.1 pmol of each). The DNA substrate (0.1 pmol, lane 1) was incubated for 1 h at 25oC with Pol 4 (0.4 pmol, lanes 2–10) in the absence (lanes 2, 5 and 8) and presence of; lane 3, 0.4 pmol full length Nej1 (Nej1F); lane 4, 0.8 pmol full length Nej1; lane 6, 0.4 pmol Nej1 N-terminal fragment (Nej1N); lane 7, 0.8 pmol Nej1 N-terminal fragment; lane 9, 0.4 pmol Nej1 C-terminal fragment (Nej1C); lane 10, 0.8 pmol Nej1 C-terminal fragment. (B) Linear DNA duplex with an internal 1-nucleotide gap was end-labeled as indicated. The DNA substrate (1 pmol, lane1) was incubated for 1 h at 25°C with Pol4 (0.4 pmol, lanes 2–10) in the absence (lanes 2, 5 and 8) and presence of; lane 3, 0.4 pmol full length Nej1 (Nej1F); lane 4, 0.8 pmol full length Nej1; lane 6, 0.4 pmol Nej1 N-terminal fragment (Nej1N); lane 7, 0.8 pmol Nej1 N-terminal fragment; lane 9, 0.4 pmol Nej1 C-terminal fragment (Nej1C); lane 10, 0.8 pmol Nej1 C-terminal fragment. After separation by denaturing gel electrophoresis, labeled oligonucleotides in the gel were detected by PhosphorImager analysis. The boxes in Panel A indicate the grouping of images from different parts of the same gel. The positions of the substrate and extended products are indicated on the left. The results of three independent experiments are shown graphically with error bars indicating the standard deviation from the mean.
