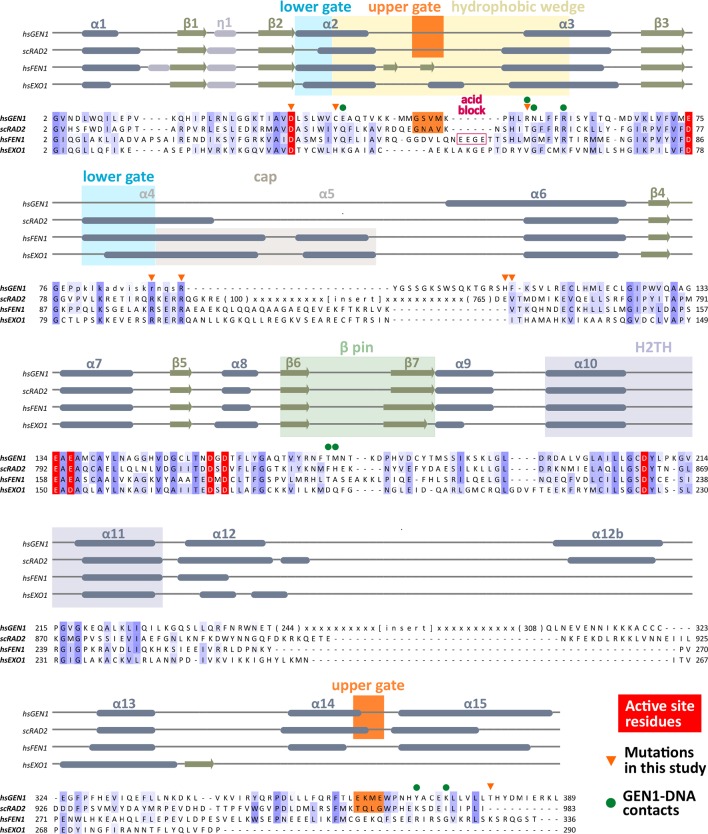
Figure 2. Alignment of the nuclease cores of Rad2/XPG-family proteins.
The alignment is based on known crystal structures: human GEN1 (PDB 5t9j, this study), yeast Rad2 (PDB 4q0w), human FEN1 (PDB 3q8k), human EXO1 (3qe9). Secondary structure elements are depicted on top of the sequence with dark blue bars for α-helices, light blue bars for 310-helices and green arrows for β-sheets. The numbering follows a unified scheme for the superfamily. Functional elements are labeled and described in the main text. Sequences are colored by similarity (BLOSUM62 score) and active site residues are marked in red. Mutations analyzed in this study are marked with an orange triangle and DNA contacts found in the human GEN1–HJ structure have a dark green dot. Disordered or missing parts in the structures are labeled in small letters or with x.