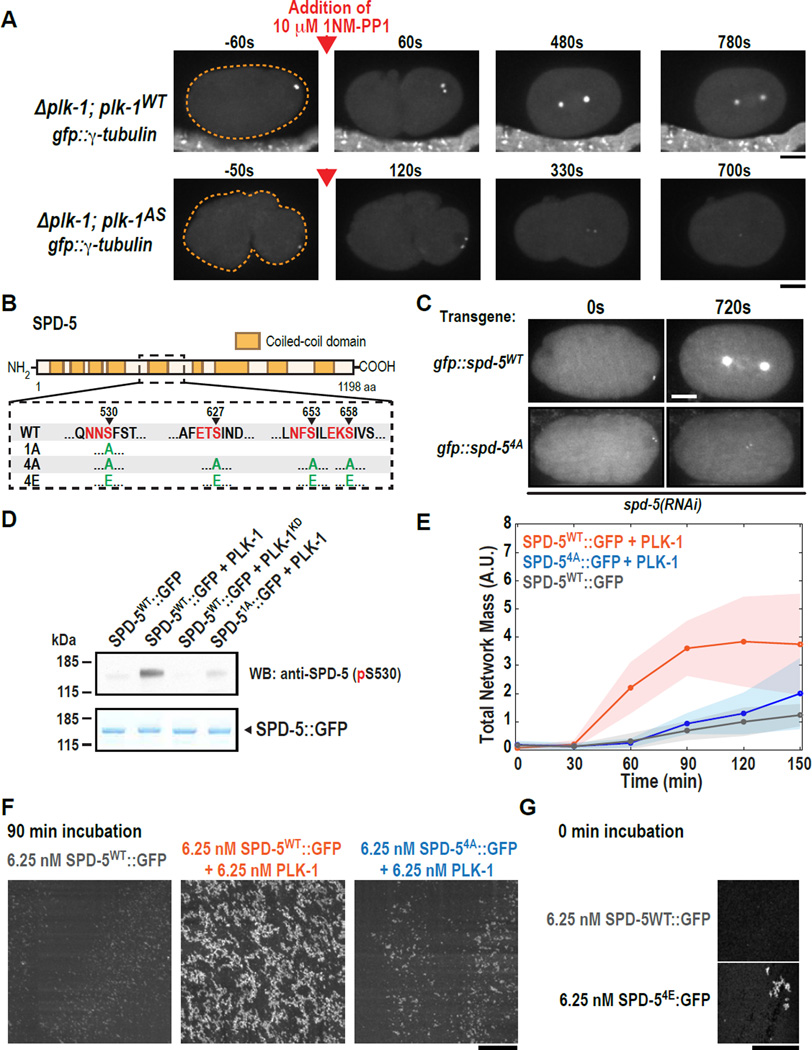
Fig. 3. PLK-1 phosphorylation of SPD-5 drives PCM assembly in vivo and SPD-5 polymerization in vitro.
(A) plk-1WT (n=9) and plk-1AS (n=13) embryos expressing the PCM marker GFP::γ-tubulin were visualized by fluorescence confocal microscopy (orange dashed line is embryo outline). 10 µM 1-NM-PP1 (PLK-1AS inhibitor) was added to permeabilized embryos prior to mitotic entry (red arrow).
(B) Diagram of phospho-epitopes on SPD-5 and different mutant constructs. Canonical PLK-1 consensus motifs (32, 33) are indicated in red. The arrowheads indicate the phosphorylated residue in each motif. The complete set of phosphorylation sites identified by MS/MS is included in Table S3.
(C) Centrosome size was visualized in embryos expressing RNAi-resistant GFP::SPD-5WT or GFP::SPD-54A. Images are sum intensity projections from z-stacks. Scale bar, 25 µm. See also Fig. S3.
(D) In vitro kinase assay. SPD-5WT::GFP was incubated with buffer alone, PLK-1WT, or a kinase dead version of PLK-1 (PLK-1KD). Phosphorylation at serine 530 was detected by western blot using a phospho-specific antibody.
(E) Kinetics of 6.25 nM SPD-5WT::GFP network formation in vitro. Values represent mean with 95% confidence intervals (n= 8–14).
(F) Representative images of SPD-5::GFP networks from (E). Scale bar, 25 µm.
(G) A phosphomitetic version of SPD-5 (SPD-54E::GFP) already formed networks at t = 0 min, bypassing the need for incubation at room temperature. Scale bar, 25 µm.