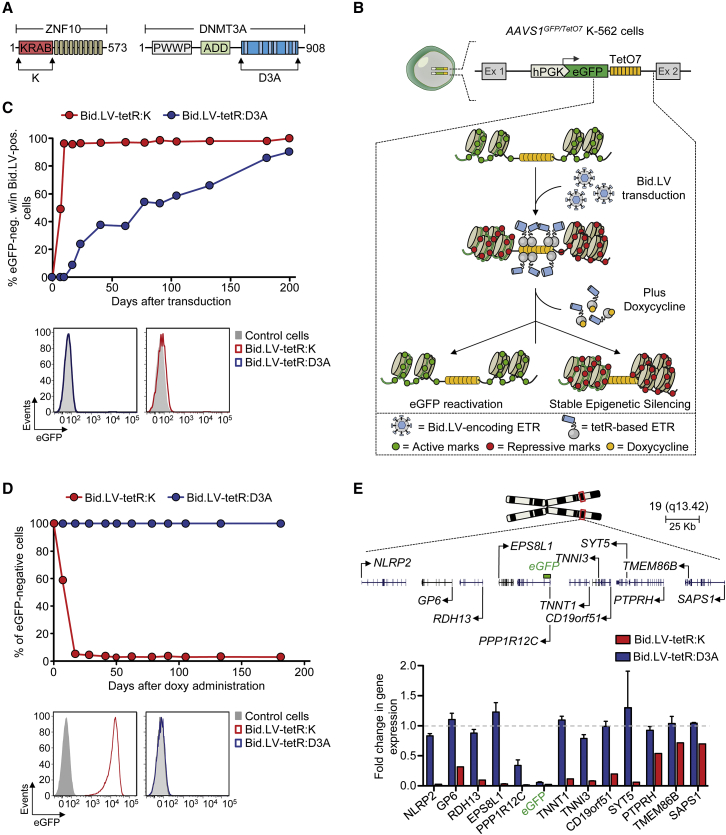
Figure 1.
Activity of the KRAB- and DNMT3A-Based ETRs
(A) Schematics of the ZNF10 and DNMT3A proteins indicating the KRAB (K) and the catalytic domain of DNMT3A (D3A).
(B) Experimental cell model used to assess activity of candidate effector domains. Top drawing shows a K-562 cell clone containing bi-allelic insertion of the hPGK-eGFP.TetO7 cassette into intron 1 of the PPP1R12C gene (a.k.a. AAVS1). The boxed bottom drawing shows the epigenetic state of the indicated region in the following experimental conditions: (1) in untreated cells, in which the region is decorated by active epigenetic marks and eGFP is expressed; (2) upon transduction with a Bid.LV expressing a tetR-based ETR, whose binding to the TetO7 element leads to deposition of repressive epigenetic marks and silencing of the cassette; (3) and upon conditional release (by doxy administration) of the ETR from the TetO7 element. In this setting, the repressive marks previously deposed by the ETR can be either erased or propagated to the cell progeny by the endogenous cell machinery, thereby leading to transcriptional reactivation or permanent silencing of eGFP expression, respectively. hPGK, human phosphoglycerate kinase gene promoter.
(C) Top: graph showing the percentage of eGFP-negative cells within the indicated Bid.LV-transduced cell populations cultured without doxy. Data are represented as mean of AAVS1GFP/TetO7 K-562 cell clones #10 and #27 of Figure S1D. Bottom: representative flow cytometry histograms of the indicated cell populations at termination of the experiment.
(D) Top: silenced cells from (C) were sorted and cultured with doxy. The graph shows the percentage of eGFP-negative cells over time. Bottom: histograms of the indicated cell populations at termination of the experiment.
(E) Top: schematic of chromosome 19 and zoom on the AAVS1 locus containing the eGFP-expression cassette. Bottom: gene expression profile of the AAVS1 locus from eGFP-negative cells transduced with the indicated Bid.LVs. The expression level of each gene was normalized to B2M and represented as fold change over a matched, untransduced AAVS1GFP/TetO7 K-562 cell clone (mean ± SEM for Bid.LV-tetR:D3A, n = 3 independent analyses; mean value for Bid.LV-tetR:K, n = 2 independent analyses). See also Figure S1 and Tables S1 and S2.