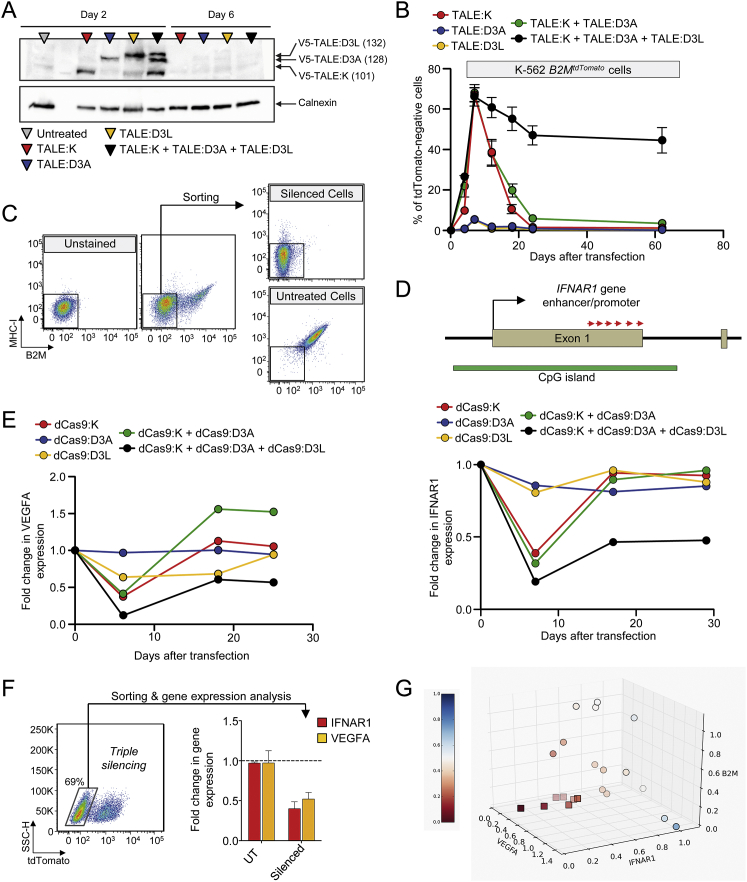
Figure S5.
Stable Silencing of Three Human Endogenous Genes by the Triple ETRs Combination, Related to Figure 4
(A) Western blot analysis of K-562 cells 2 (left) or 6 (right) days upon transfection with plasmids encoding for the indicated V5-tagged TALE-based ETRs. Blots were probed with an anti-V5 tag or anti-Calnexin antibody, the latter used as loading control. The expected molecular weight (in KDa) of each protein is indicated.
(B) Time-course flow cytometry analysis of B2MtdTomato K-562 cells upon transfection with plasmids encoding for the indicated TALE-based ETRs. Data show percentage of tdTomato-negative cells (mean ± SEM; n = 3 independent transfections for each treatment condition).
(C) Representative dot plots showing the sorting strategy used to obtain B2M-negative HEK-293T cells (top right) from bulk-treated, ETR-silenced cells (middle). Unstained cells are shown (left).
(D) Top: schematic of the IFNAR1 promoter/enhancer region showing the orientation of the sgRNAs (red arrows) and the CGI. Bottom: Graph showing the kinetics of IFNAR1 silencing (measured as fold change in mRNA levels over untreated cells, HPRT1 as normalizer) in K-562 cells transfected with plasmids encoding for the indicated dCas9-based ETRs and a pool of 6 sgRNAs targeting the IFNAR1 promoter/enhancer region.
(E) Graph showing the kinetics of VEGFA silencing (measured as fold change in mRNA levels over untreated cells, HPRT1 as normalizer) in K-562 cells transfected with plasmids encoding for the indicated dCas9-based ETRs and a pool of 12 sgRNAs targeting the VEGFA promoter/enhancer region.
(F) Representative dot plot of B2MtdTomato K-562 cells treated for multiplex gene silencing (left) and expression analysis of the indicated genes from tdTomato-negative cells (right). The expression level of the indicated genes was normalized to HPRT1 (mean ± SEM of 2 independent transfections for each treatment condition). Analysis at 25 days post-transfection.
(G) Three-dimensional scatterplot depicting the expression levels of B2M, IFNAR1 and VEGFA in 21 K-562 cell clones derived from a multiplex gene silencing experiment performed with dCas9-based ETRs and a pool of sgRNAs targeting the three genes. Axes represent the fold change in mRNA levels of the indicated genes over a mean of three untreated cell populations (HPRT1 as normalizer). Clones with consistent and concurrent downregulation of the three genes are identified by squares. Grouping was performed with Density-Based Spatial Clustering of Applications with Noise (DBSCAN). Points are colored by their euclidean distance from origin of the cartesian space, this corresponding to the ideal perfect triple silencing (0,0,0). Transparency reflects distance from the observer, solid marks are the closest.