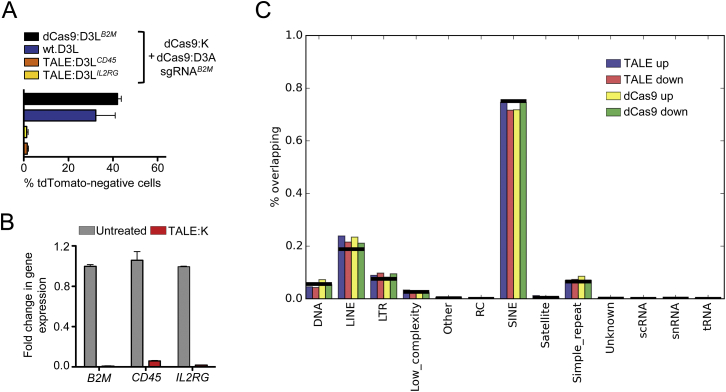
Figure S7.
Specificity Analyses of the Triple ETR Combination, Related to Figure 7
(A) Histogram showing the percentage of tdTomato-negative cells at 25 days post-transfection with plasmids encoding for the indicated TALE- and dCas9- based ETRs with or without the wild-type DNMT3L (wt.D3L). Data are shown as mean ± SEM of 3 independent transfections for each treatment condition.
(B) Histogram showing the fold change in the expression levels of the indicated genes at day 7 upon transient transfection of plasmids encoding for TALE:K targeting the indicated genes. Data are represented as mean ± SEM of 3 independent transfections for each treatment condition.
(C) Enrichment of DMRs (defined at low stringency cutoff: nominal p value < 0.01, logFC > 1) in repetitive elements, defined by RepeatMasker annotation from UCSC Genome Browser track. For each experiment, the ratio of selected DMRs overlapping a specific class of repetitive element was calculated. Horizontal black bars represent the ratio of all analyzable methylated regions (n = 198886) over repetitive elements. None of the ratios was found significantly higher than the expected (test: chi-square).