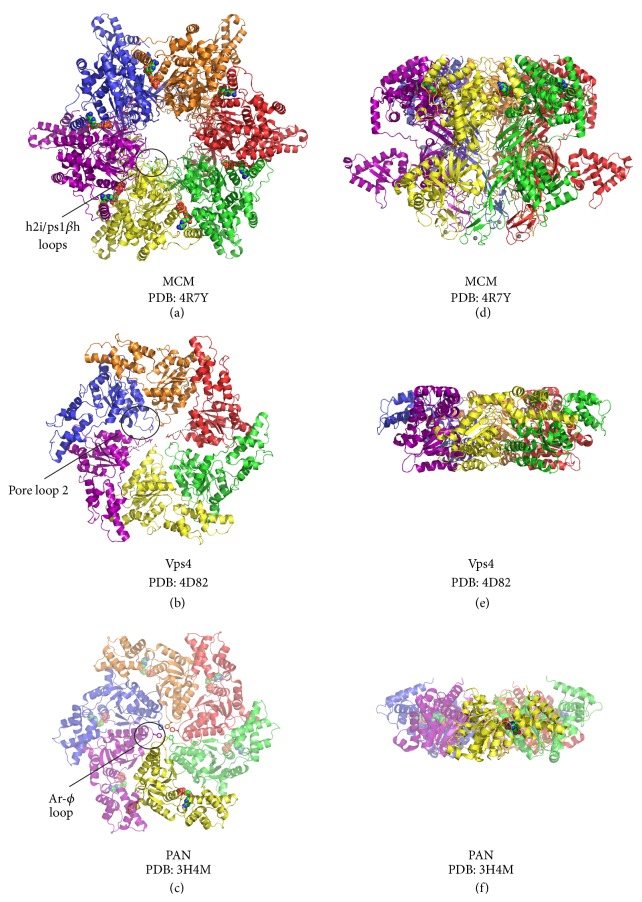
Figure 3.
Shared structural features of archaeal AAA+ hexameric translocases. Views of MCM, Vps4, and PAN crystal structures are shown parallel (a–c) and perpendicular (d–f) to the central channel with each subunit uniquely colored and in cartoon representation. In the views parallel to the central channel, the C-terminal face of each complex is projected out of the page. In the perpendicular view, the C-terminus is located at the top and the N-terminus is located on the bottom. (c, f) The PAN hexamer is a model of the nucleotidase ring generated by superimposition of six copies of the PAN monomer atomic coordinates onto the six subunits of of the HslU hexamer (PDB: 1DO0) [49, 50]. Five of the ADP-bound PAN monomers used to generate the model of a hexamer are shown in transparent view (c, f). DNA or protein substrate interaction motifs are projected into the central channel with representative pore loops circled and labelled (a–c). Highlighted pore loop residues are shown in stick, magnesium ions as magenta spheres (a, d), ADP molecules in space-filling view (a, d, c, f), and zinc ions as gray spheres (a, d). All structure representations in the figure were prepared with the Pymol software package [43] and PDB accession codes 4R7Y [41] (a, d), 4D82 [46] (b, e), and 3H4M [50] (c, f).