Abstract
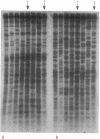
Recombination is unknown in natural populations of Rivulus marmoratus, a selfing hermaphrodite, and genetic variation is likely due to mutation alone. DNA fingerprinting with an array of microsatellite [e.g., (CT)9] and minisatellite (e.g., the 33.15 core sequence) probes reveals very high clonal diversity within samples of seven Floridian populations, of which five contain about as many clones as there are individuals. There are 42 clones among 58 individuals surveyed (mean, 1.4 individuals per clone), a level of genetic diversity unprecedented among clonal animals. Moreover, all of the probes recognize the same clones even though, at high hybridization stringencies, there is little overlap in the fingerprint patterns they generate. This suggests that most sympatric clones differ by multiple and independent mutational steps. In one population studied in detail, the average number of mutational steps separating two clones is estimated at 9 or 10 and may be substantially higher. The mutational discontinuities among sympatric clones make it unlikely that they evolved by accumulation of neutral mutations in populations that are otherwise genetically uniform. The data argue that the mixing of unrelated individuals from different local populations occurs to an extent previously unappreciated and/or that divergence of clones is mediated by natural selection. If confirmed, the latter would be a serious challenge to current ideas on the predominant role of recombination in promoting the evolution of biological novelty.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atz J. W. Hermaphroditic Fish. Science. 1965 Nov 5;150(3697):789–797. doi: 10.1126/science.150.3697.789. [DOI] [PubMed] [Google Scholar]
- Caskey C. T., Pizzuti A., Fu Y. H., Fenwick R. G., Jr, Nelson D. L. Triplet repeat mutations in human disease. Science. 1992 May 8;256(5058):784–789. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- Chandley A. C., Mitchell A. R. Hypervariable minisatellite regions are sites for crossing-over at meiosis in man. Cytogenet Cell Genet. 1988;48(3):152–155. doi: 10.1159/000132613. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974 Oct;78(2):737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh J. The accumulation of deleterious genes in a population--Muller's Ratchet. Theor Popul Biol. 1978 Oct;14(2):251–267. doi: 10.1016/0040-5809(78)90027-8. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Royle N. J., Wilson V., Wong Z. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature. 1988 Mar 17;332(6161):278–281. doi: 10.1038/332278a0. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Ledwith B. J., Manam S., Nichols W. W., Bradley M. O. Preparation of synthetic tandem-repetitive probes for DNA fingerprinting. Biotechniques. 1990 Aug;9(2):149–152. [PubMed] [Google Scholar]
- Lynch M. The similarity index and DNA fingerprinting. Mol Biol Evol. 1990 Sep;7(5):478–484. doi: 10.1093/oxfordjournals.molbev.a040620. [DOI] [PubMed] [Google Scholar]
- Mitani K., Takahashi Y., Kominami R. A GGCAGG motif in minisatellites affecting their germline instability. J Biol Chem. 1990 Sep 5;265(25):15203–15210. [PubMed] [Google Scholar]
- Shin H. S., Bargiello T. A., Clark B. T., Jackson F. R., Young M. W. An unusual coding sequence from a Drosophila clock gene is conserved in vertebrates. Nature. 1985 Oct 3;317(6036):445–448. doi: 10.1038/317445a0. [DOI] [PubMed] [Google Scholar]
- Turner B. J., Elder J. F., Jr, Laughlin T. F., Davis W. P. Genetic variation in clonal vertebrates detected by simple-sequence DNA fingerprinting. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5653–5657. doi: 10.1073/pnas.87.15.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]