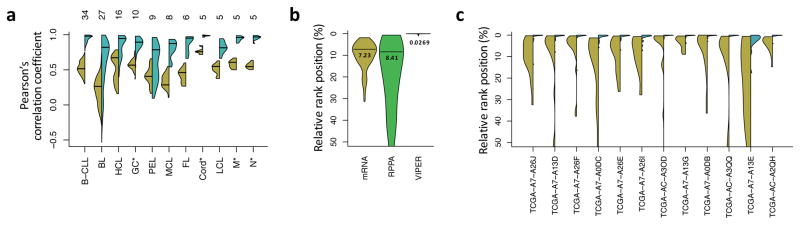
Figure 3. Reproducibility of VIPER results.
(a) Violin plot showing the distribution of correlation coefficients computed between all possible pairs of gene expression signatures (yellow) or VIPER protein activity signatures (cyan) for samples of the same B cell phenotype, including normal (GC, germinal center reaction; M, memmory and N, peripheral blood B cell) and pathologic (B-CLL, B cell chronic lymphocytic leukemia; BL, Burkitt lymphoma; HCL, hairy cell leukemia; PEL, primary effusion lymphoma; MCL, mantle cell lymphoma; FL, follicular lymphoma) phenotypes. The number of samples per phenotype is indicated on top of the figure. (b) Probability density for the relative rank position of the most upregulated gene (mRNA, yellow), relatively abundant protein (RPPA, green) or activated protein (VIPER, cyan), identified in each profiled basal breast carcinoma sample, across all the remaining profiled samples. The horizontal line and number beneath indicates the distribution mode. (c) Probability density for the relative rank position of the top 10 most upregulated genes (yellow) or VIPER-inferred activated proteins (cyan), identified from FF samples on the corresponding FFPE samples.