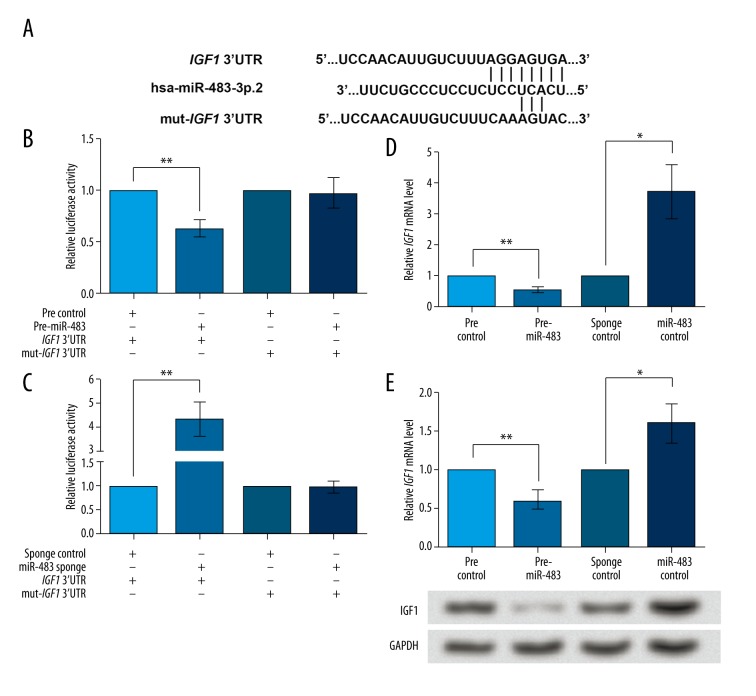
Figure 3.
IGF1 is directly inhibited by miR-483. pre-miR-483, KGN cells transfected with pre-miR-483. miR-483 sponge, KGN cells transfected with miR-483 sponge. pre/sponge control, KGN cells transfected with the corresponding blank vector as a control. (A) The sequence AGGAGUGA in IGF1 3′UTR is predicted to be the binding site of miR-483. Five bases in the binding site are mutated to construct mut-IGF1 3′UTR. (B) Luciferase reporter assay shows that the activity of wildtype IGF1 3′UTR, but not mut-IGF1 3′UTR, can be inhibited by miR-483 overexpression compared to the control. (C) Luciferase reporter assay shows that the activity of wildtype IGF1 3′UTR, but not mut-IGF1 3′UTR, can be up-regulated by miR-483 sponge compared to sponge control. (D) IGF1 mRNA expression was inhibited by miR-483 overexpression and promoted by miR-483 sponge, as shown by qRT-PCR. (E) IGF1 protein level was inhibited by miR-483 overexpression and promoted by miR-483 sponge, as shown by Western blot. GAPDH is used as an internal reference. * P<0.05. ** P<0.01. IGF1, insulin-like growth factor1. CCNB1 – cyclin B1. CCND1 – cyclin D1. CDK2 – cyclin-dependent kinase 2.