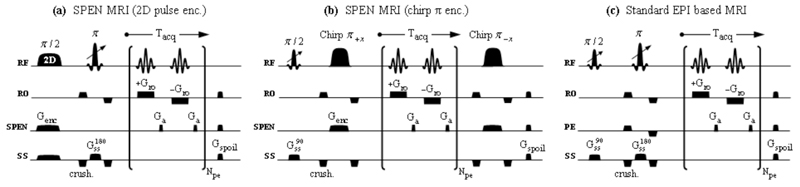
Figure 1.
Three-dimensional multi-slice imaging sequences assayed in this work, based on acquiring series of single-scan 2D images for different z-coordinate values. (a) SPEN-based sequence employing a two-dimensional excitation RF pulse acting to spatiotemporally-encode one of the dimensions, and a slice-selective π pulse with variable offset (indicated by the arrow) to select the orthogonal one. (b) Alternative encoding approach employing a standard 1D π/2 slice-selective excitation and a non-specific spatiotemporal encoding π-pulse (chirped adiabatic passage). Full gradient and RF rewindings are applied following each acquisition block, allowing the use of short inter-scan recycle delays and minimizing the residual T1 weighting of subsequently acquired slices. (c) EPI-based sequence employing slice-selective excitations and spin-echo pulses, followed by a standard k-space 2D encoding block. Spoiler gradients were added to all three sequences in order to dephase any residual transverse magnetizations that may have been left between the acquisitions of consecutive slices. In all cases the 2D data matrices were collected in a single scan per slice-encoding increment. Axes definitions are as summarized in Table 1; additional abbreviations used include exc: excitation, crush: crusher gradients, SPEN: spatiotemporal encoding channel, SS/ss: slice-selective channel, RO/ro: readout channel, PE/pe: phase-encoded channel, acq: acquisition, Npe: number of phase-/SPEN-encoded lines.