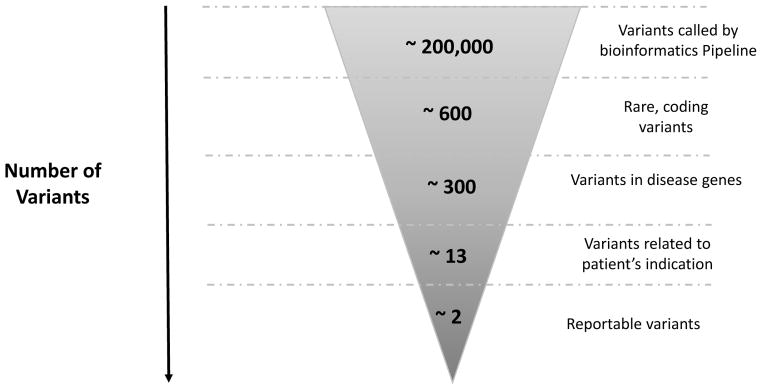
Figure 2.
Number of variants at each step in the clinical exome bioinformatics filtration pipeline and interpretation process. As shown, ~200,000 variants are called by the bioinformatics pipeline following alignment of the captured sequence to a reference genome. Initial filtration is to select rare variants affecting exons ± 6bp. Variants that are high frequency in the general population (and therefore likely to be polymorphisms) are removed by filtering using population databases, primarily ExAC. Variants that are known to be disease causing are identified by filtering against databases with known mutations such as the Human Gene Mutation Database (HGMD). After this, on average, ~600 variants are retained, of which ~300 are in known disease genes. Subsequently, the remaining variants are manually correlated with the patient phenotype to identify genes that may be associated with the patient’s clinical features. An average of 13 variants remain, of which ~2 variants are likely to be clinically significant and therefore reportable (These are either pathogenic, likely pathogenic variants or variants of uncertain clinical significance). Incidental findings in the 56 ACMG gene list are reported in ~5% of cases. Data was derived from 182 clinical exome sequencing tests performed at the Division of Genomic Diagnostics, Children’s Hospital of Philadelphia (approximate averages are represented).