FIGURE 6.
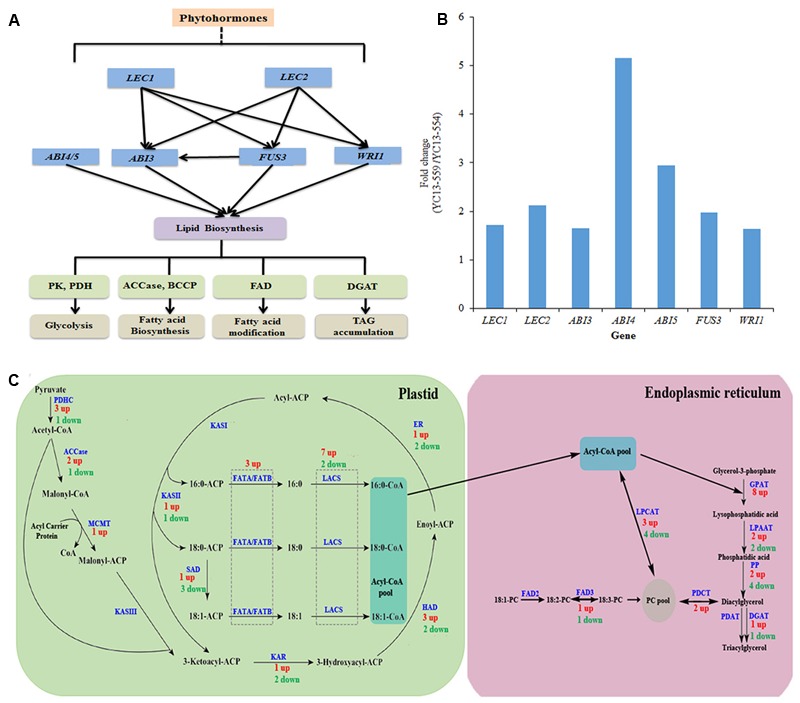
Overview of gene network involved in seed oil biosynthesis in B. napus(A). A simplified scheme of transcription factors (TF) network involved in regulation seed oil biosynthesis B. napus (B) Relative expression levels of different TF genes in YC13-559. TFs whose expression changed more than 1.5-fold in YC13-559 relative to YC13-554 are shown. The RPKM values for multiple copies of same TF were summed. (C) Overview of de novo fatty acid (FA) and triacylglycerol (TAG) biosynthesis pathways. Genes whose expression changed more than 1.5-fold in YC13-559 relative to YC13-554 were included in this study. Genes were mapped onto de novo FA and TAG biosynthesis pathway. Numbers refers to the numbers of genes, and “up” refers to up-regulation in YC13-559 line compare to YC13-554 line, and “down” to down-regulation. Same enzyme catalyze several steps are highlighted with dotted rectangle. Lipid substrates are abbreviated: 16:0, palmitic acid; 18:0, stearic acid. PDHC, pyruvate dehydrogenase complex; ACCase, acetyl-CoA carboxylase; MCMT, malonyl-CoA : ACP malonyltransferase; ACP, acyl carrier protein; KASI/II/III, ketoacyl-ACP synthase I/II/III; KAR, ketoacyl-ACP reductase; HAD, hydroxyacyl-ACP dehydrase; ER, enoyl-ACP reductase; FATA/B, fatty acyl-ACP thioesterase A/B; SAD, stearoyl-ACP desaturase; LACS, long-chain acyl-CoA synthetase; GPAT, glycerol-3-phosphate acyltransferase; LPAAT, 1-acylglycerol-3-phosphate acyltransferase, PP, phosphatidate phosphatase; PDCT, Phosphatidylcholine:diacylglycerol cholinephosphotransferase; PDAT, phospholipid :diacylglycerol acyltransferase; DGAT, acyl-CoA : diacylglycerol acyltransferase, LPCAT, lysophospholipid acyltransferase; FAD2/3, fatty acid desaturase.
