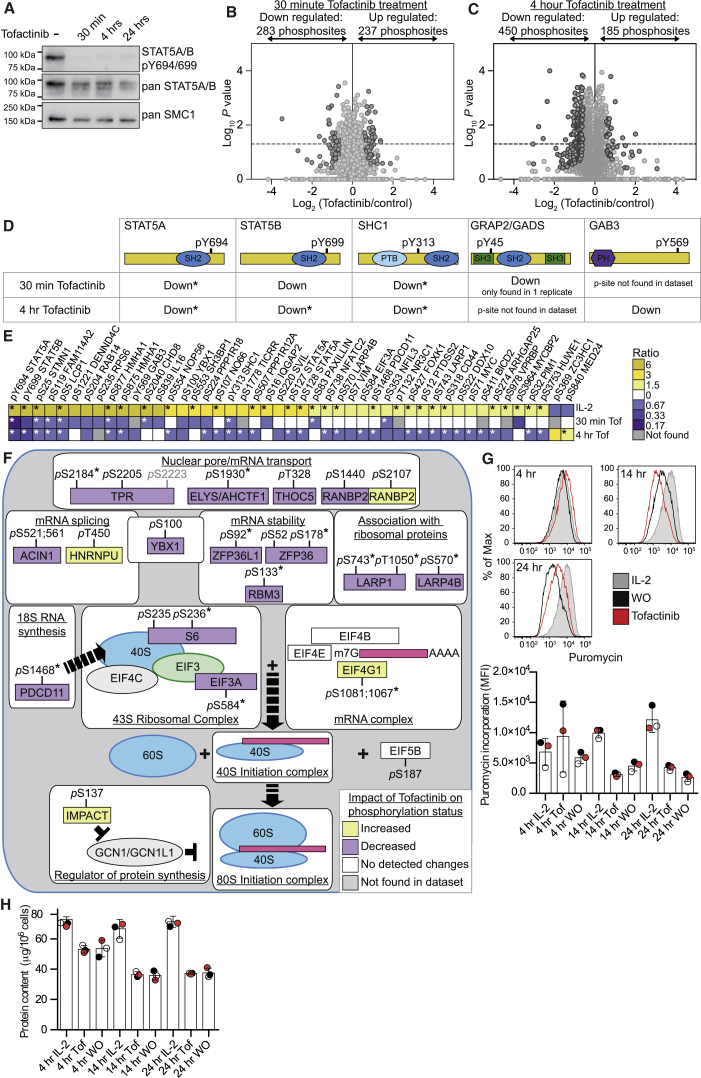
Figure 3.
Analysis of the Impact of the JAK1/JAK3 Inhibitor, Tofacitinib, on the IL-2-Maintained CTL Phosphoproteome
(A) Immunoblot analysis of the STAT5 phosphorylation in CTLs maintained in IL-2 only over a time course of treatment with 100 nM Tofacitinib, a JAK1/3 inhibitor.
(B and C) Heavy-labeled SILAC CTLs were treated with 100 nM Tofacitinib for 30 min (B) or 4 hr (C) before being mixed with control (light) CTLs for phosphoproteome analysis. Log-transformed SILAC ratios are plotted against log-transformed p values (one sample t test). See also Figure S1, and refer to Tables S4 and S5 for lists of phosphopeptides. The graphs in (B) and (C) show the phosphosites identified in three biological replicates. Phosphosites with ratios reproducibly changed by 1.5-fold are shown in dark gray.
(D) The impact of Tofacitinib on the IL-2-regulated Tyr phosphorylation sites is shown.
(E) The heatmap shows phosphosites inversely regulated by IL-2 and Tofacitinib.
(F) Tofacitinib-regulated phosphorylation sites identified in proteins involved in mRNA processing, transport, stability, and translation are shown. See also Table S6.
(D–F) Phosphosites found to show a statistically significant regulation (p value ≤ 0.05, one sample t test) are marked with an asterisk (∗).
(G) Protein synthesis of CTLs maintained in IL-2 only, or IL-2 maintained CTL treated with 100 nM Tofacitinib (Tof) or deprived of any cytokines (WO) for different times was measured by puromycin incorporation using a flow-cytometry-based assay. Representative flow cytometry plots of the puromycin incorporation measurements are shown, with color-matched individual replicate data from three biological replicates shown alongside. The quantified protein content, as determined by BCA assay, of the same cells at each time point is shown in (H). Bars in (G) and (H) show the mean ± SD.