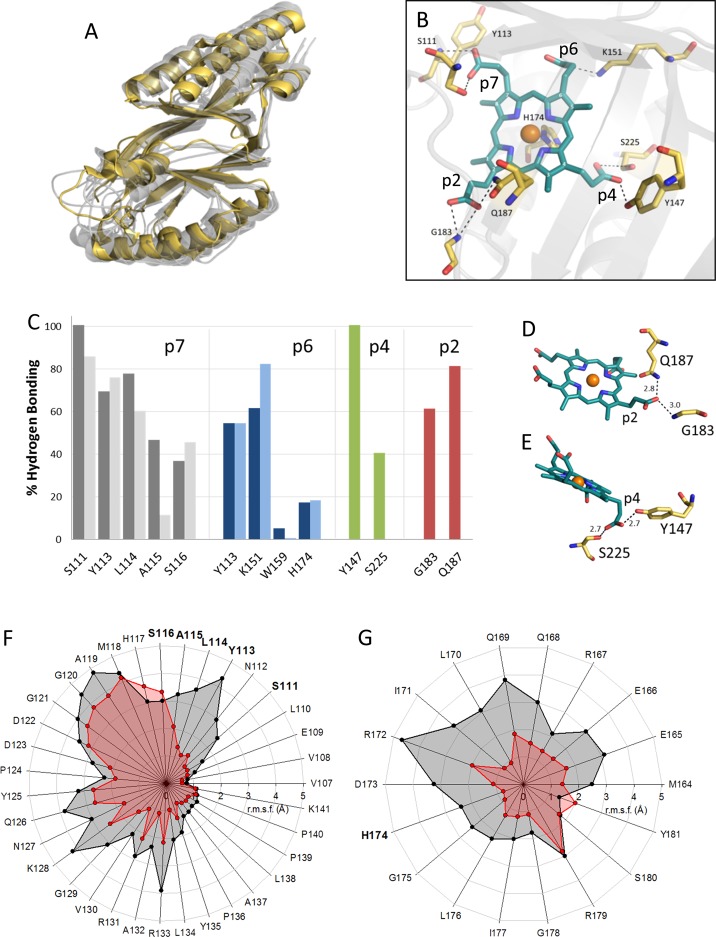
Figure 8.
Proposed interactions of coproheme with HemQ based on MD simulations. (A) Overlay of subunit structures of apo-LmHemQ (yellow, PDB entry 4WWS) and coproheme-LmHemQ (gray, chains A–E) from MD simulations (last frames). (B) Hydrogen bonding network of coproheme in LmHemQ (last frame, chain A), in which positions of porphyrin substituents are numbered in white. (C) H-Bonding interaction of atoms of respective amino acids and coproheme propionates (for details, see Table S1) and for heme b propionates. H-Bonds formed (in percent) throughout the simulation time as averages over all five subunits are represented as dark (coproheme) and light (heme b) gray bars (residues H-bonded to propionate at position 7), dark (coproheme) and light (heme b) blue bars (p6), red bars (p4), and green bars (p2). H-Bonding of (D) p2 to coproheme (last frame, chain A) and (E) p4 to coproheme (last frame, chain C). (F) Root-mean-square fluctuations (in angstroms) for all atoms (averages over all five chains) of the flexible loop on the entrance to the coproheme/heme b binding site between residues V107 and K141 in apo-LmHemQ (black) and coproheme-LmHemQ (red) over 30 ns simulations. (G) Root-mean-square fluctuation analysis as in panel F for the α-helix (M164–Y181) that harbors the proximal histidine (H174).