Fig. 3.
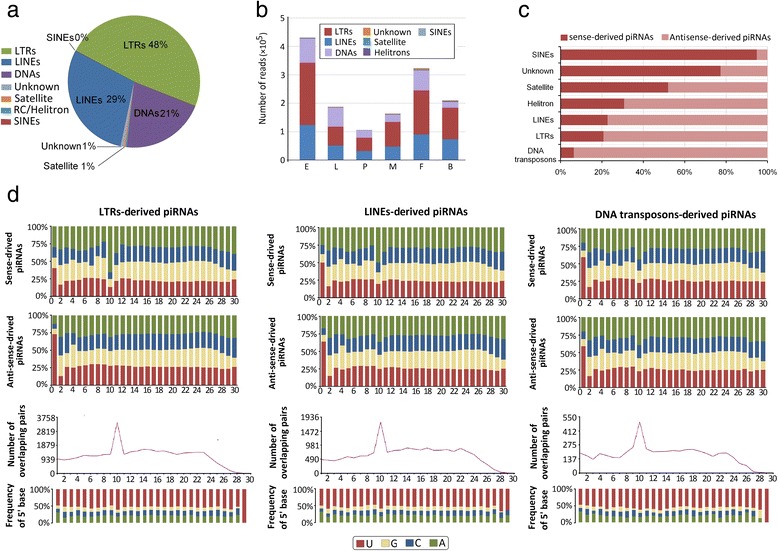
Characterization of repeat-derived piRNAs. a The proportion of piRNAs mapped to repeat sequences shows that major piRNAs are preferentially produced from LTR, LINE, and DNA transposons within the transposon group. b Abundance and distribution of repeat-derived piRNAs in various developmental stages; repeat-derived piRNAs displayed similar compositions. c Strand preference of repeat-derived piRNAs. d Upper panel: base composition of repeat-derived piRNAs of three major TE sequences, LTR, LINE, and DNA transposons. The X-axis represents the nucleotide position relative to the 5′ ends of the piRNAs. The Y-axis represents the percentage of base bias. Lower pane: ping-pong pair analysis of three major TE sequences, LTR, LINE, and DNA transposons. The length of overlap is shown on the horizontal axes. Indicated above each axis is the number of possible overlapping pairs of small RNAs with the specified overlap size. Indicated below each axis is the relative frequency of the 5′ base identity for overlapping sequences. The colour code for the bases is indicated in the centre box
