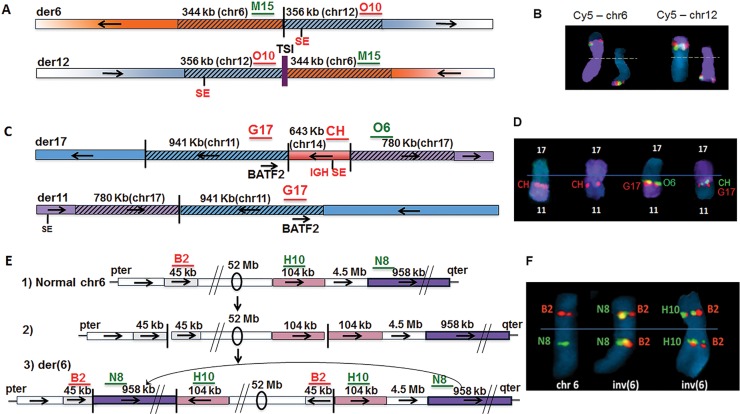
Figure 3.
Complex interchromosomal non-MYC locus rearrangements in three MMCL. (A) t(6;12) with duplications on both derivative chromosomes and insertion of chr1 TSI on der(12) in KP6 MMCL; (B) Metaphase FISH of t(6;12) in KP6, with localization of chr6 probe (M15, green signal) and chr12 probe (O10, red signal) on der(6) and der (12), with chr6 and chr12WCP probes (purple), respectively, on the left and right; (C) t(11;17) with duplicated sequences from chr11 (including BATF2) and chr17 breakpoints at der11 and der17 breakpoint junctions, and insertion of chr14 segment including 3′ IGH SE on der17 in MOLP8 MMCL; (D) Metaphase FISH showing localization of probes on der17; data not shown for der11; (E) A total of 52 Mb chr6 inversion with duplicated breakpoint sequences, and insertion of distant sequences from 6q at the 6p breakpoint junction in JJN3 MMCL: (1) anatomy of normal chr6, with B2, N8, H10 indicating the approximate position of FISH probes; (2) hypothetical inversion precursor without insertion; (3) observed inversion with insertion; (F) Metaphase FISH showing chr6 and inv(6) with probes for relevant sequences (B2, H10, N8). Probe details in Supplementary Table S5. Horizontal arrows indicate orientation of chromosomal segments.