Figure 1.
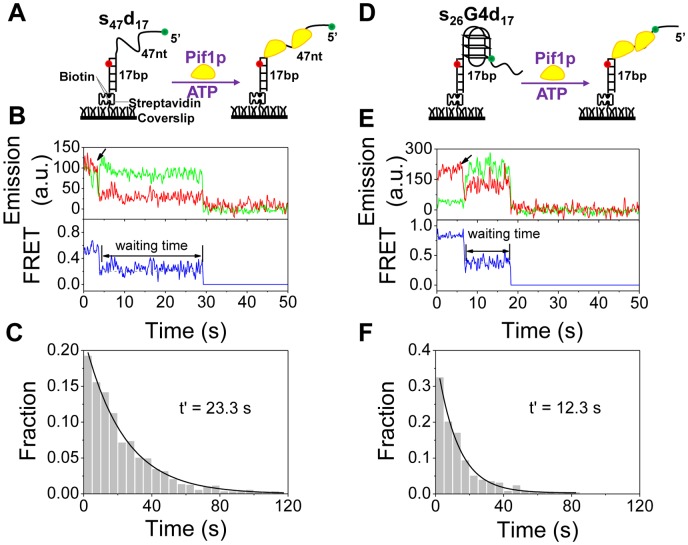
G4 structure reduces the waiting time for downstream duplex DNA unwinding by Pif1p. (A) Schematic representation of experimental procedures for partial duplex DNA unwinding. (B) Representative fluorescence emission and FRET traces for s47d17 unwinding by 80 nM Pif1p and 2 mM ATP. Upon addition of Pif1p and ATP as indicated by the arrow, FRET signal decreases abruptly, indicating the binding of Pif1p. It then remains at a constant value for ∼25 s and then the Cy3 and Cy5 signals disappear simultaneously as the duplex is separated. The waiting time is defined as the time between Pif1p binding and duplex separation. (C) Histogram of the waiting time follows an exponential decay with a time constant of 23.3 s. (D) Schematic representation of experimental procedures for G4/ds DNA unwinding. (E) Representative fluorescence emission and FRET traces for Pif1p-catalyzed unwinding of s26G4d17. Upon Pif1p and ATP addition as indicated by the arrow, FRET signal decreases rapidly, then keeps at constant value for ∼12 s, until both Cy3 and Cy5 signals disappear abruptly. (F) Histogram of the waiting time follows an exponential decay with a time constant of 12.3 s.