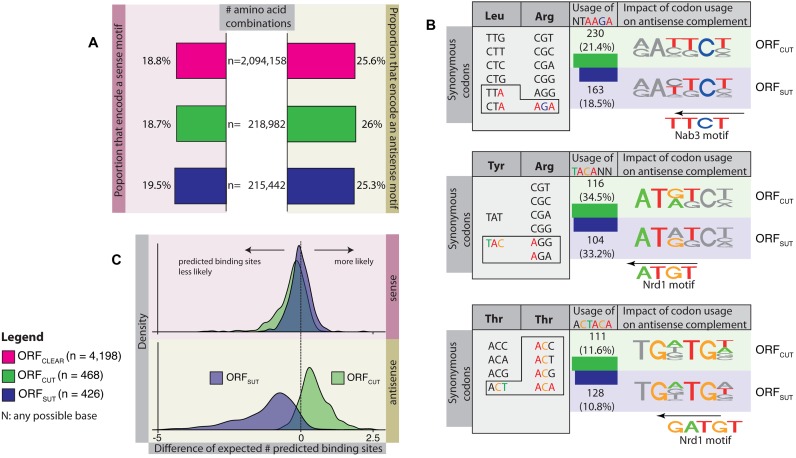
Figure 2.
Comparison of amino acid and codon usage in different ORF types. (A) Bar plots showing the proportions of amino-acid pairs and triplets that encode Nrd1 or Nab3 motifs in the sense and antisense directions. There is no difference in amino-acid pair and triplet content in the different ORF types. (B) Examples of variable codon usage for amino acid pairs in different ORF types. Bar plots show the use of codon combinations in ORFSUT and ORFCUT that encode a binding motif. Although amino acid content is the same across all ORF types, there is a small difference in codon pair usage that affects the occurrence of predicted antisense binding sites. ORFCUT tend to use more codon pairs encoding a motif than ORFSUT. (C) Distributions of the expected numbers of predicted binding sites in ORFCUT and ORFSUT given the underlying codon usage normalised by the expected numbers of predicted sites given the background codon usage for ORFCLEAR. Positive values indicate a larger number of predicted sites than expected (given the background usage), and vice versa for negative values. The distributions for ORFSUT and ORFCUT in the sense direction are centred on 0 (mean = −0.28 and s.d. = 0.5 for ORFCUT; mean = 0.00 and s.d. = 0.39 for ORFSUT), indicating similar numbers of sites as ORFCLEAR. The antisense distribution shows that ORFSUT favour a codon usage pattern that yields fewer predicted binding sites than ORFCLEAR or ORFCUT (mean = 0.48 and s.d. = 0.56 for ORFCUT; and mean = −1.15 and s.d. = 1.07 for ORFSUT).