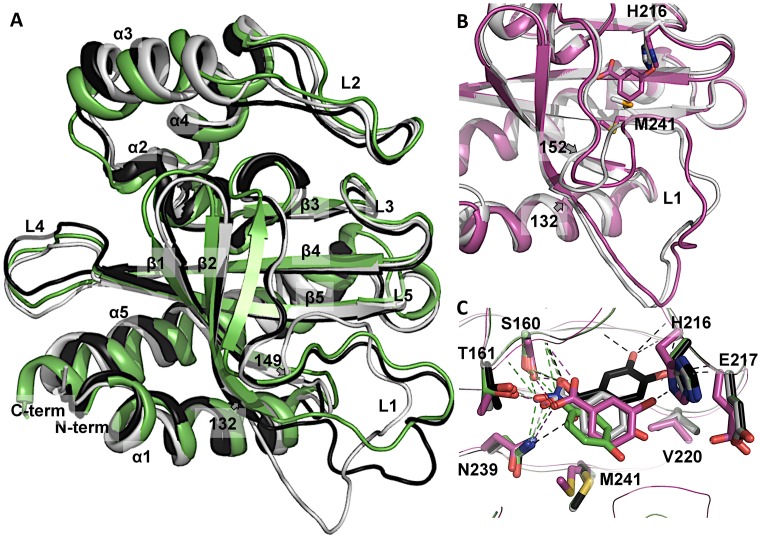
Figure 2.
DoubleMut-IBD homology model and crystal structure. (A) Monomer of DoubleMut-IBD crystal structure (light gray) overlaid on DoubleMut homology model (black) and the template (PDB code: 2IA2, green). (B) Overlay of apo- (light gray) and holo- (magenta) crystal structures of DoubleMut, showing conformational changes in H216 and M241 side chains and rearrangement of loop L1 (residues 132–152). (C) DoubleMut-3HB crystal structure (magenta) overlaid on pre-determined ligand-docked homology models of DoubleMut with 4HB (light gray), 34DHB (black) and pNP (green). Carboxylate and nitro group of ligands showing polar interaction with backbone of S160, and side chains of T161 and N239 are highlighted with dotted lines. Figures were created using PyMOL (Version 0.99, Schrödinger, LLC).