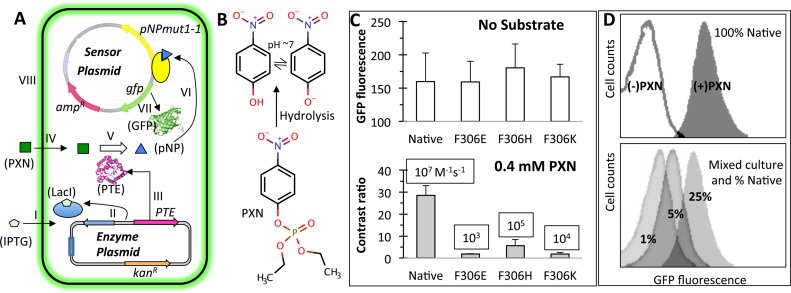
Figure 4.
‘Smart’ microbial cells that hydrolyze paraoxon (PXN) and sense the product pNP. (A) Two plasmid system consisting of an Escherichia coli cell with a ‘sensor plasmid’ and an ‘enzyme plasmid’. Functioning of a smart cell is shown in steps I–VIII. IPTG induced expression of a phosphotriesterase (PTE) enzyme (steps I-III) hydrolyses exogenously supplied PXN substrate (steps IV–V), releasing pNP that activates the engineered TF, pNPmut1-1 (step VI) to express the reporter gene, gfp (step VII). Intracellular green fluorescent protein accumulation results in fluorescence of the cell (step VIII). The figure has been derived from our previous publication (26). (B) Substrate PXN and hydrolysis product pNP. (C) PTE variants (Native, F306E, F306H and F306K) expressed in ‘smart’ cells in the absence of substrate (top panel) and presence of 0.4 mM PXN (bottom panel, showing contrast ratio). Published catalytic efficiency of each PTE variant (30) is displayed in a box. Error bars represent standard deviation from two independent experiments performed on different days and divalent metal concentration between 0.2 and 0.5 mM. (D) Fluorescence histograms of ‘smart’ cells expressing native PTE and grown in the absence and presence of PXN (top panel). Fluorescence histograms of a mix of population with Native % as indicated and F306E, F306H and F306K in equal proportion, grown to an OD (600 nm) of 0.6 and then spiked with IPTG and PXN (bottom panel).