Figure 1. Identification of novel HLA-G regulatory miRs by in silico based approach.
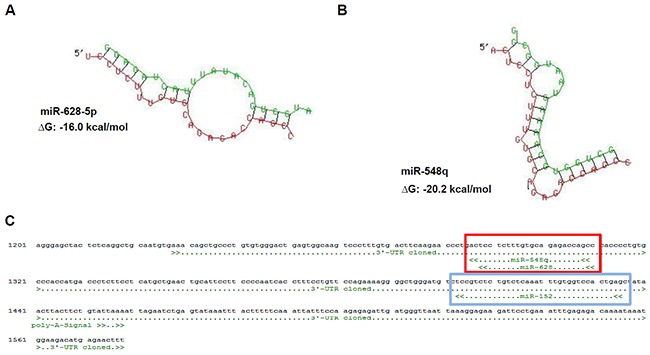
A, B. Sequence alignment and prediction of secondary structure including a calculated free energy for the HLA-G 3′-UTR (red) and the novel identified HLA-G regulatory miRs (green) miR-628-5p (A) and miR-548q (B) by usage of the free online data base RNAhybrid [32]. C. In silico predicted binding sites of the novel identified miRs miR-548q and miR-628-5p (red box), which share the same position at the HLA-G 3′-UTR and is in the 3′ direction localized next to the binding site of the miR-148 family (e.g. miR-152; blue box, [14]).
