Figure 2. Characterization of the direct interaction between miR-548q, miR-628-5p and the HLA-G 3′-UTR.
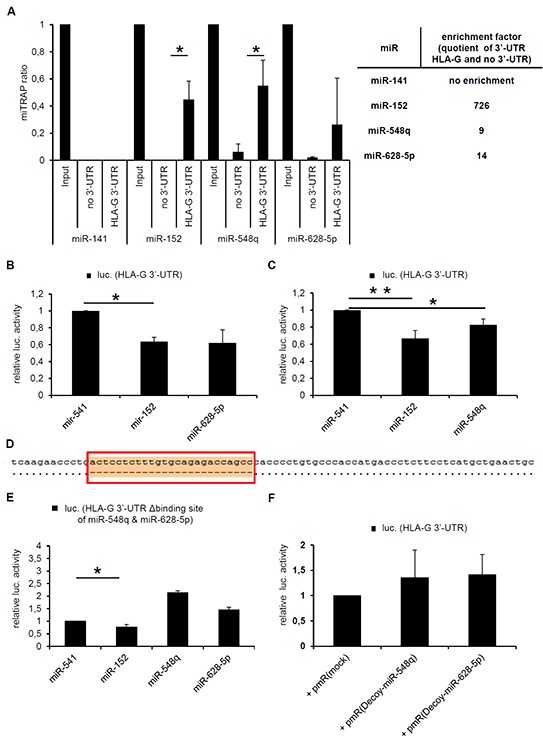
A. The results of three miTRAP experiments using in vitro transcribed MS2 loop-tagged HLA-G 3′-UTR as bait as described in Materials and Methods followed by qPCR analysis of the miTRAP eluates are shown. The results are expressed as miTRAP ratio [31]. Furthermore, a quotient (enrichment factor) of specifically enriched miRs (HLA-G 3′-UTR as bait) and the non-specifically enriched miRs (mock sequence as bait) was calculated and summarized in the adjacent table. The published HLA-G-regulatory miR-152 served as positive control and showed a higher enrichment with the HLA-G 3′-UTR when compared to the miR-548q and miR-628-5p. The HLA-G non-regulatory miR-141 was expressed in the cell lysate, but not enriched with the HLA-G 3′-UTR. B, C. The direct interaction of the miRs miR-548q and miR-628-5p with the HLA-G 3′-UTR was further investigated by luc reporter gene assays as described in Materials and Methods. The results were normalized to the overexpressed HLA-G non-relevant control miR-541, which is not expressed in the applied HEK293T cells and is only reported to play a role in neuronal differentiation [51]. The overexpression of miR-152 served as positive control. In comparison to the negative control miR-541 the overexpression of miR-152, -628-5p and -548q downregulated the luc reporter gene activity of the reporter gene construct encoding the HLA-G 3′-UTR. However, the effect of miR-152 is much stronger than the effect of miR-628-5p or miR-548q. The results were normalized to the overexpressed HLA-G non-relevant control miR-541. D. Sequence alignment of the HLA-G wt 3′-UTR (pubmed data base; NM_002127.5) with the HLA-G 3′-UTR (G*010103) including the deletion of the binding site of miR-548q and miR-628 (red box) is visualized after sequencing. The binding site of miR-152 remains unaffected. E. No downregulation of the luc reporter gene activity was observed upon a deletion of the binding site for miR-628-5p and miR-548q in the HLA-G 3′-UTR by overexpression of miR-548q and miR-628-5p. Overexpression of miR-152 still down regulates the luc reporter gene activity due to its intact binding site. F. A luc reporter gene assay in combination with the wt HLA-G 3′-UTR is shown. Instead of miR expression vectors so called decoy constructs against the miR-548q and miR-628-5p present in the HEK293T cells were transfected, after Haraguchi et al., 2009 [52] leading to a non significant stabilization of the luc reporter gene activity when compared to the respective mock vector.
