Figure 2.
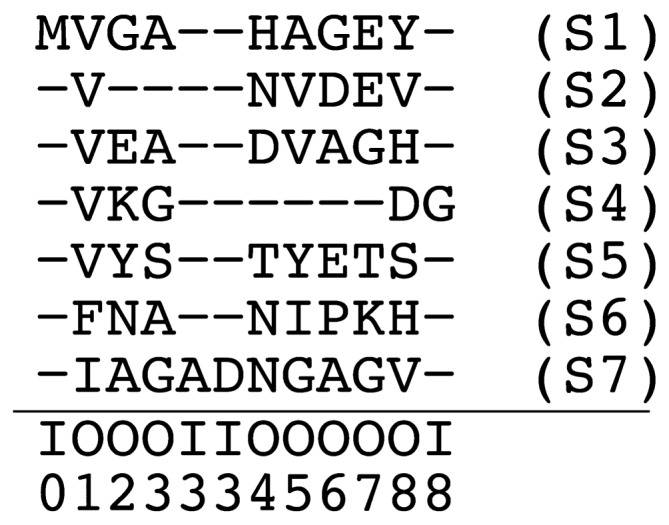
Example of a multiple sequence alignment (based on [1]). Each row corresponds to a protein sequence (S1,…,S7) and each column to an alignment site. Below the horizontal line, each alignment site is annotated as to whether it corresponds to a core (matching or deletion) site (“O”) or an insert site (“I”). Indicated below these “O”/”I” symbols are the position of lattice sites. (c.f. Fig. 1) The size of the lattice model based on this MSA is N=8. Insert sites other than I0, I3 and I8 are not explicit in this MSA. For example, the alignment of the sequence S2 in this figure is represented as XS2=X0…X9=(O0,−)(O1,V) (O2,−)(O3,−)(O4,N)(O5,V)(O6,D)(O7,E)(O8,V)(O9,−) where the first and last pairs represent the start and end of the alignment, respectively. As another example, the alignment of sequence S7 is XS7=X0 … X11=(O0,−) (O1,I)(O2,A)(O3,G)(I3,A)(I3,D)(O4,N)(O5,G) (O6,A)(O7,G)(O8,V)(O9,−).
