Abstract
Metabolic pathway reconstruction presents a challenge for understanding metabolic pathways in organisms of interest. Different strategies, i.e., reference-based vs. de novo, must be used for pathway reconstruction depending on the availability of well-characterized enzymatic reactions. If at least one enzyme is already known to catalyze a reaction, its amino acid sequence can be used as a reference for identifying homologous enzymes in the genome of an organism of interest. Where there is no known enzyme able to catalyze a corresponding reaction, however, the reaction and the corresponding enzyme must be predicted de novo from chemical transformations of the putative substrate-product pair. This review summarizes studies involving reference-based and de novo metabolic pathway reconstruction and discusses the importance of the classification and structure-function relationships of enzymes.
Keywords: enzyme, chemical transformation, substrate-product pair, genome, metabolome
Natural products serve as important sources of drugs [1] and provide insight into ecological factors, such as inter-species relationships [2,3]. Within the plant kingdom, estimates on the number of metabolites have ranged from 200,000 [4,5] to 1,060,000 [6], while the number of human metabolites is believed to be over 40,000 [7]. Thus, across all of nature, the number of metabolites is likely to far exceed these values. Identification of these metabolites and their associated metabolic pathways has the potential to provide significant benefits, not only in terms of pharmaceuticals and the public health, but also in terms of agricultural and environmental issues. However, the number of enzymes that have been verified experimentally and approved by the International Union of Biochemistry and Molecular Biology (IUBMB) is limited to approximately 5,600 [8], and the total number of reactions associated with those enzymes is approximately 8,100 [9]. Although experimentally verified metabolites and enzymatic reactions are not directly comparable in number, this large discrepancy reveals that our knowledge of metabolites and metabolic pathways is limited to only a small portion of all natural products.
Experimental identification of metabolites is difficult, expensive, and time consuming. Recent advances in gas chromatography-mass spectrometry [10], liquid chromatography-mass spectrometry (LC-MS) [11,12], capillary electrophoresis-MS [13,14], and nuclear magnetic resonance (NMR) spectroscopy [15] have enabled rapid and comprehensive analysis of numerous metabolites. However, analyzing the entire metabolome as part of a systems biological approach remains impractical [16]. In particular, it is difficult to structurally assess intermediates that are present at low concentrations. The experimental identification, purification, and characterization of enzymes also present challenges [17].
These challenges necessitate the use of in silico techniques for predicting the chemical structures of intermediate compounds, putative reactions between compounds, and the enzymes responsible for reactions as comprehensively as possible. In this review, we provide an overview of recent in silico studies that have contributed to the field of metabolic pathway reconstruction.
Classification of enzyme functions
The naming of enzymes is essential for the reconstruction of metabolic pathways. Challenges arise when the same name is given to different enzymes or the same enzyme is given more than one name, and both situations should be avoided where possible. For this purpose, some attempts have been made to classify enzymes according to their functions. The first such attempt was based on the number of substrates and products involved in the reaction [18], classifying enzymes into three reaction types: (1) A+B=C+D, (2) A=B+C, and (3) A=B. However, this classification system did not become widespread.
The second system for enzyme classification was developed in 1958 and classified 659 enzymes into hydrolyzing enzymes, transferring enzymes, and others [19]. This system was later improved, with enzymes being sorted into six classes according to the type of reaction, and became the current classification scheme used for the Enzyme List of the Nomenclature Committee of IUBMB (NC-IUBMB) [20]. In this system, each enzyme is given a unique four-digit code, the Enzyme Commission (EC) number, in which the first, second, and third digits represent a hierarchical classification of enzymes (referred to as class, subclass, and sub-subclass, respectively) (Fig. 1). The third edition of the NC-IUBMB Enzyme List was printed in 1992, comprising over 850 pages. Further printed versions were deemed unfeasible, not only because of the large number of pages required, but also because the data quickly become out of date. This led to an online version, ExplorEnz [8], which is the primary repository for all enzymes classified by the IUBMB. ExplorEnz accepts proposals for the addition of new enzymes or the modification of existing entries. Newly approved enzymes are regularly made public at http://www.enzyme-database.org/newenz.php.
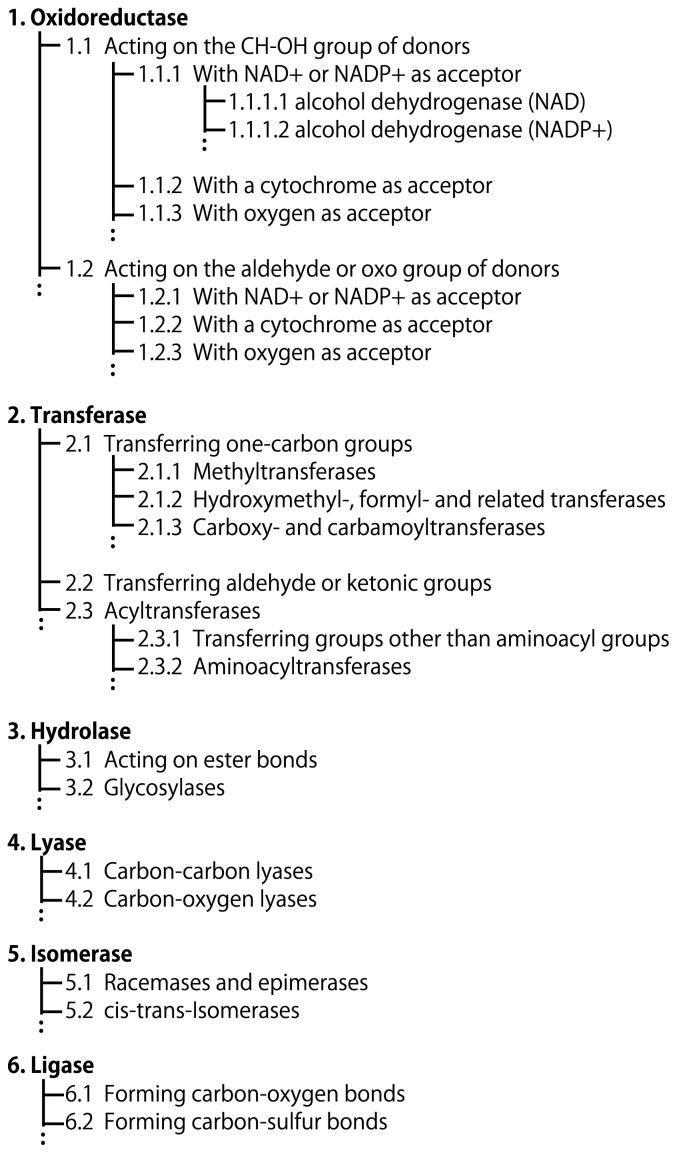
Figure 1.
Hierarchical classifications of enzymes by the NC-IUBMB Enzyme List. Enzymes are given IDs, namely the Enzyme Commission (EC) numbers, consisting of four digits connected by dots (e.g., EC 1.1.1.1). The first digit represents one of the six classes, 1. oxidoreductases, 2. transferases, 3. hydrolases, 4. lyases, 5. isomerases, and 6. ligases. The second and third digits represent more detailed classification. The classification criteria are different dependent on the classes.
Reference-based metabolic pathway reconstruction
Collective use of the Enzyme List enables the reconstruction of metabolic pathways. However, missing reactions are an issue for several reasons. First, the Enzyme List only contains enzymes that have been fully characterized experimentally. Because it is difficult to automatically update the list, some enzymes may have already been reported but may not be listed yet. Second, some enzymes may catalyze alternative reactions, but not all substrates and products may be listed exhaustively in the Enzyme List. Third, spontaneous reactions are not present in the Enzyme List.
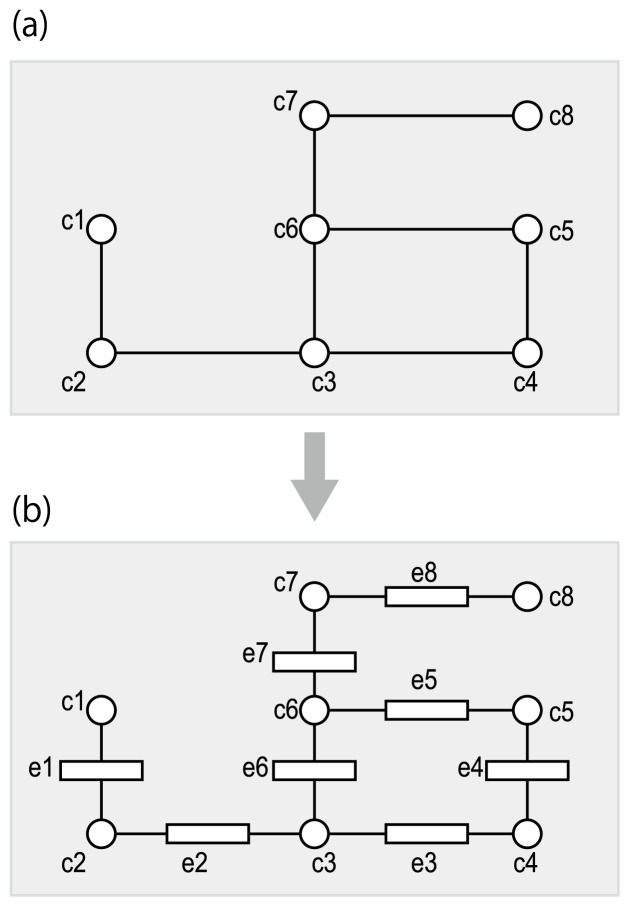
Many reaction databases (such as BRENDA [21], KEGG [9], MetaCyc [22], and Rhea [23]) use the Enzyme List to reconstruct metabolic pathways and supplement with additional reactions to fill in the gaps. These reconstructed pathways can be represented as combined pathways that only describe chemical transformations without distinguishing between organisms (Fig. 2). For example, the glycolytic and tricarboxylic acid (TCA) cycle pathways are present in human cells, but the gluconeogenesis pathway is not. Nevertheless, these three pathways share some common compounds and thus can be combined when we only consider chemical transformations. Such combined pathways are referred to as “reference pathways” [24].
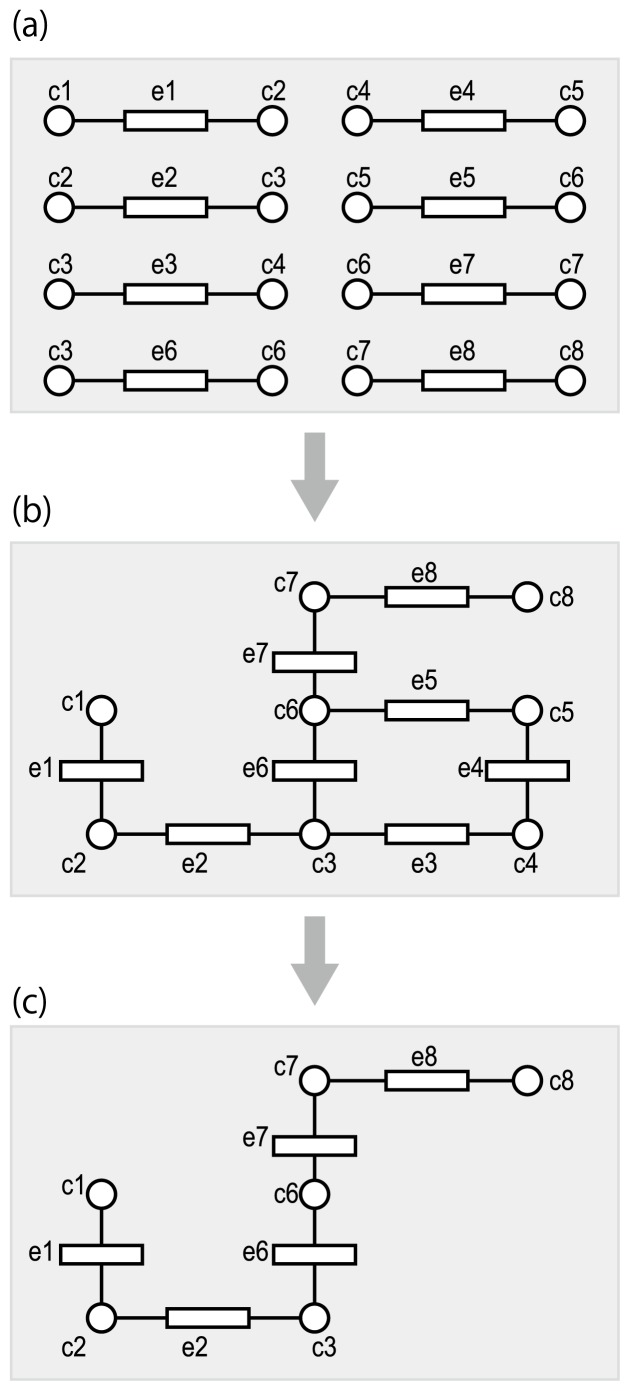
Figure 2.
Reference-based metabolic pathway reconstruction. Circles and rectangles represent metabolic compounds (metabolites) and enzymes, respectively. (a) Substrate-product relationships in enzymatic reactions, which can be derived from the NC-IUBMB Enzyme List. (b) The reference pathway is reconstructed by connecting reactions with common metabolites. The reference pathway only concerns chemical transformations of metabolites, and does not take into account any differences in specific organisms. In other words, the reference pathway is a combined pathway taken from many organisms. (c) An organism-specific pathway is derived by mapping putative enzymes in the genome of interest. In order to achieve this goal, it is necessary to find known enzyme proteins that are similar to the putative enzyme proteins.
These reference pathways are useful when comparing metabolic models in different organisms (Fig. 2(c)). Most (but unfortunately not all) EC numbers can be associated with the amino acid sequences of the responsible enzyme proteins. Putative enzyme genes may be predicted based on orthology, i.e., homologous sequences descended from a common ancestral sequence. In other words, EC numbers may be predicted on the basis of amino acid sequence similarity. With a comprehensive set of putative enzyme genes in an organism of interest, genes can be assigned to their appropriate positions in the pre-defined reference pathways based on orthology [25–34].
There have been some attempts at automated reference-based metabolic pathway reconstruction at the genome-wide scale. For example, MetaCyc provides reconstructed pathway from BioCyc, a collection of organism-specific pathways [22], and the KEGG database provides organism-specific pathways for complete genomes. Their reconstruction processes are based on ortholog assignments derived from best hits to the complete genomes of various organisms using sequence similarity search programs such as BLAST, followed by manual curation [9,22]. KEGG also provides an automated web-server named KEGG Automatic Annotation Server (KAAS: http://www.genome.jp/tools/kaas/), enabling reference-based metabolic pathway reconstruction on demand [35]. The model SEED, which uses a table-like representation of functionally related enzyme genes called a subsystem, automates reconstruction using a completed genome sequence [36]. More recently, improved tools with greater efficiency and interpretability have been developed, including MG-RAST [37] and MEGAN [38] for pathway reconstruction and analysis of species distributions in large metagenomic data, MAPLE [39] for easy interpretation of the availability of metabolic functions, and BlastKOALA and GhostKOALA [40] for efficient ortholog assignments using reduced sets of reference genome data. These programs can be used for gaining insight into the metabolic potential of various environments.
An increasing number of new genome-scale metabolic models have been reconstructed, indicating the value of reference-based metabolic pathway reconstruction [41]. Experimental validation remains necessary to ensure the validity of the metabolic models, and improvement of the models enables more precise prediction of physiological characteristics. This in silico approach represents an attempt at assigning well-defined reactions to putative enzyme genes (or enzyme proteins), i.e., predicting EC numbers from sequence similarity. High sequence similarity alone, however, is not thought to be sufficient to assign an EC number, as a minor sequence change may alter enzyme activity or specificity (e.g., stilbene synthase and chalcone synthase [42]) and enzyme proteins may be bifunctional (e.g., ribulose 1,5-bisphosphate carboxylase/oxygenase [43] and luciferase [44]). Thus, incorporation of other types of evidence is necessary to reconstruct more accurate metabolic models, such as gene orders, phylogenetic profiles, and gene expression profiles [32,33,45–50].
De novo metabolic pathway reconstruction
Reconstructed pathways are inherently incomplete due to the lack of experimentally identified enzymes and metabolites. There may be gaps (missing reactions) between known reactions even in central metabolic pathways. These missing reactions may lead to erroneous interpretations during metabolic analysis. In addition, most natural products are produced in only a subset of organisms. Such natural product pathways are mostly unknown, as previously explained. In such cases, reference-based metabolic pathway reconstruction is not possible.
This necessitates de novo metabolic pathway reconstruction, i.e., the prediction of reactions based on the chemical structures of metabolites. These methods are classified into two categories: the first predicts pathways by generating intermediate compound structures where necessary (Fig. 3), and the second uses pre-defined chemical compounds and predicts pathways by filling in the reactions between them (Fig. 4).
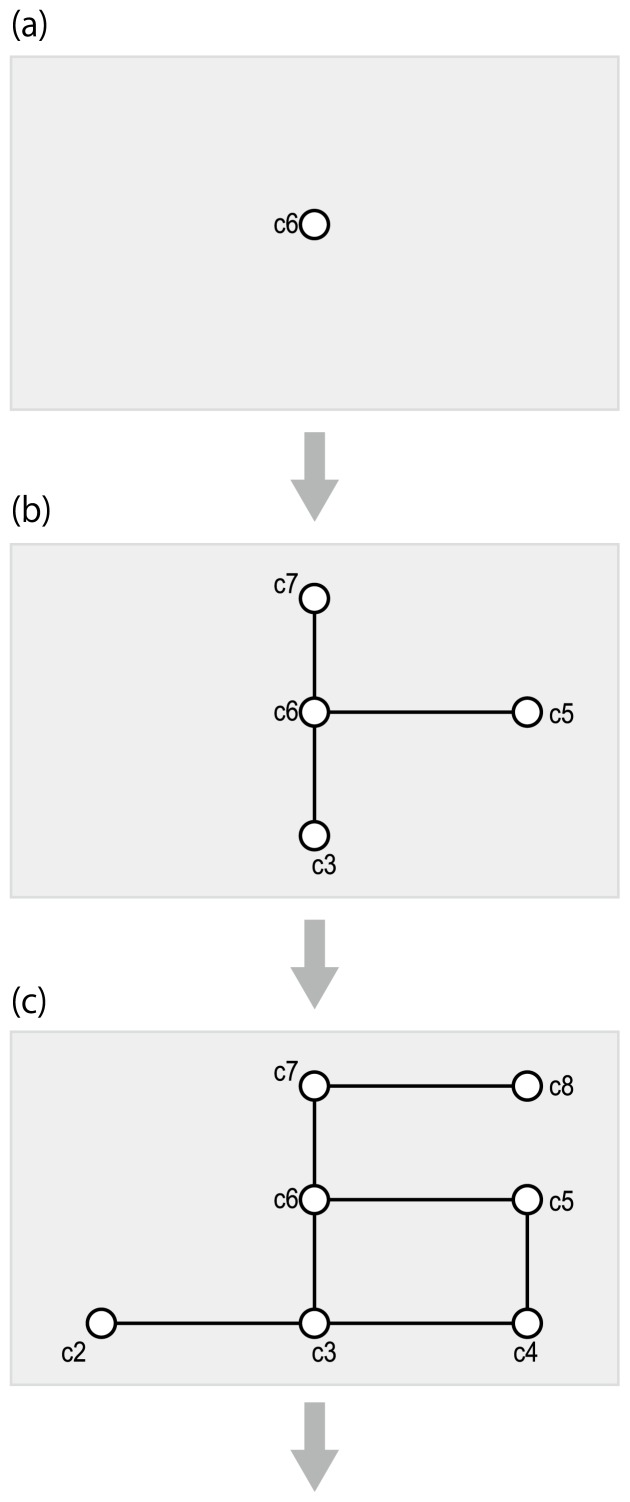
Figure 3.
Compound-filling framework of de novo metabolic pathway reconstruction. (a) The input is the chemical structure of a xenobiotic compound for a biodegradation prediction, or a final product compound for a biosynthesis prediction. (b) Pre-defined chemical transformation rules are applied so that the structures of intermediate compounds are automatically generated. (c) This process is iterated until it reaches to a known chemical compound.
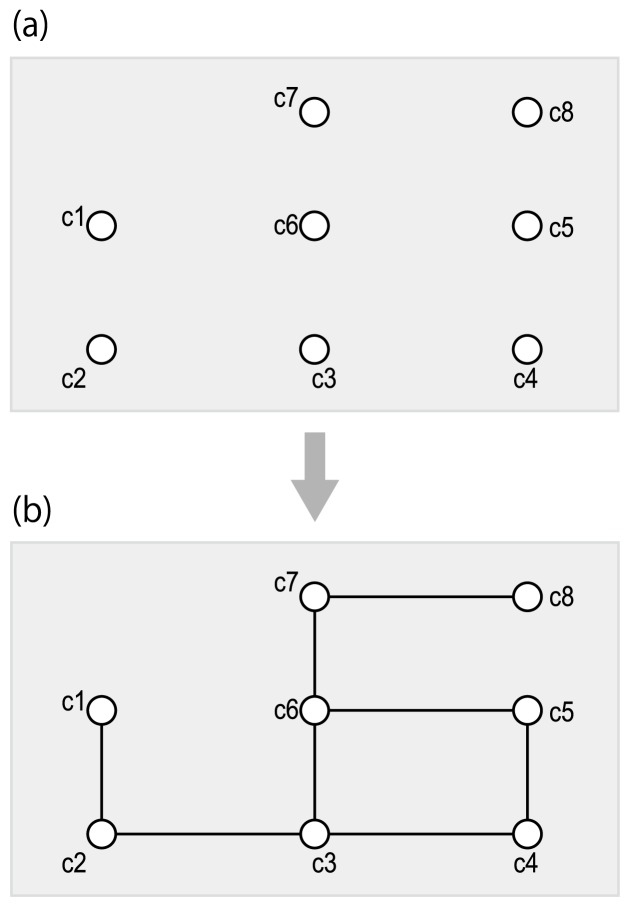
Figure 4.
Reaction-filling framework of de novo metabolic pathway reconstruction. (a) The input is a set of chemical structures of metabolites. (b) Putative reactions (that are not known yet) are predicted between the input compounds.
We refer to the first category (Fig. 3) as belonging to a “compound-filling framework”. The computer programs in this category accept a query compound and iterate the process of automatically generating the chemical structures of the next compound(s) in the predicted pathway. A similar problem has been tackled in the field of synthetic organic chemistry [51], where computer programs assist in designing strategies for synthesizing compounds of interest [52]. In both metabolic pathway reconstruction and synthetic organic chemistry, analysis is based on pre-defined chemical transformation rules, but the collection of the rules differs between the two fields. For example, carboxylic halide or carboxylic anhydride is usually used in acylation reactions in synthetic organic chemistry, whereas carboxylic thioester is mainly used in acylation reactions in metabolic pathways. Thus, the pre-defined chemical transformation rules and their prioritizations are the keys for effectively predicting pathways.
Several methods for de novo prediction of metabolic pathways using a compound-filling framework have been published [53–55], and more recently, freely available web servers have been developed [56,57]. Each defines its own chemical transformation rules that are repetitively applied to the compounds, resulting in de novo metabolic pathways. Compounds generally have more than one substructure that can undergo chemical transformation, and the number of such substructures increases as compounds become larger. Additionally, as synthetic pathways become longer, the number of possible intermediates expands in a combinatorial explosion. These methods, however, avoid such difficulties by their own design.
The Pathway Prediction System (PPS) [57] focuses on biodegradation of xenobiotic compounds in soil and provides an interactive interface that enables users to select only the reaction of interest. It also prioritizes some chemical transformation rules based on the actual activity in soil [58]. PathPred [56] deals with biosynthesis of plant secondary metabolites as well as biodegradation of xenobiotic compounds. It applies a similarity search against the KEGG database to prioritize intermediates similar to known compounds.
A common strategy of these studies is to limit the number of chemical transformation rules. It is generally known that every specific pathway, such as biodegradation of xenobiotic compounds, primarily follows its own specific chemical transformation rules [59]. This enables a computational reduction to some extent. Nevertheless, the computational costs of the compound-filling framework can become prohibitive, and it is not suitable for predicting pathways for several compounds simultaneously.
We refer to the second category (Fig. 4) of de novo pathway reconstruction as belonging to a “reaction-filling framework”, which uses many pre-defined chemical compounds and predicts pathways by filling in reactions between them. This approach is becoming more widely available owing to the availability of databases containing increasing numbers of chemical compounds for which the chemical structures have been identified. The database can be screened to search for candidate compounds with values that are obtained experimentally, such as accurate mass [60,61].
The reaction-filling methods can be classified into those that depend on pre-defined chemical transformation rules [62,63] and those that do not [64,65]. These methods face what can be regarded as a problem of enzymatic reaction-likeness, i.e., whether the given pairs of metabolites can be chemically interconverted by individual enzymatic reactions. These methods suffer from huge computational costs, and large-scale prediction is not computationally feasible.
Recently, a method was published to predict the presence/absence of enzymatic reactions between compounds; this method is computationally efficient enough to deal with tens of thousands of metabolites at a time using supervised-learning [66]. In this method, each chemical compound is represented by a chemical fingerprint (a binary vector that describes the chemical characteristics of a compound) or a chemical descriptor (an integer vector that describes the chemical characteristics of a compound) [67]. A compound pair consisting of a substrate and a product is represented by the feature vector generated from the chemical fingerprints/descriptors of the substrate and the corresponding product. The feature vector is designed to extract conserved and altered features during the putative reaction and is used for a reaction-similarity learning process based on the support vector machine (SVM).
The use of chemical fingerprints/descriptors allows for rapid searching of molecules from a vast number of molecules in a database, and this technique has been especially utilized for pharmaceutical purposes. For the purpose of metabolic pathway reconstruction in the reaction-filling framework, existing eight fingerprints (CDK fingerprint, CDK extended fingerprint, CDK graph only fingerprint, CDK hybridization fingerprint, E-state fingerprint, Klekota-Roth fingerprint, MACCS fingerprint and PubChem fingerprint) [67] were used for performance comparison, however, no significant difference was observed among them [66]. This problem may be caused by the nature of binary vectors (1 for presence or 0 for absence), which is not suited for metabolic pathway reconstruction. For example, in the case of a reaction where a carboxylate group in the substrate turns into an amide group in the product, if the substrate has only one carboxylate group and the product has only one amide group, then the feature vectors of the reaction can adequately describe the chemical change. Such binary vectors, however, can only describe the presence or absence of functional groups and not the number of such groups; thus, they are not suitable for describing reactions where the substrate has more than one carboxylate group and/or the product has more than one amide group.
A type of chemical descriptor termed KEGG Chemical Function and Substructures (KCF-S) was designed to tackle this issue and improve de novo metabolic pathway reconstruction [68]. This integer vector counts the numbers of substructures, including various functional groups. KCF-S defines substructures based on seven attributes (atom, bond, triplet, vicinity, ring, skeleton, and inorganic), imitating the terminology and process of recognizing substructures in organic chemistry or biochemistry. The most fundamental substructure attribute is referred to as the KEGG Atoms, which take physicochemical environmental properties into account and are able to discriminate between important functional groups, such as aldehyde and carboxylate. This new chemical descriptor is more efficient than previously existing fingerprints and descriptors for de novo metabolic pathway reconstruction and has exhibited clear improvements in predictive performance [68].
Another attempt to improve de novo metabolic pathway reconstruction involves a method for distinguishing regioisomers (positional isomers), information that is critical for appropriate interpretation of metabolome data [61] but cannot be distinguished by many chemical fingerprints/descriptors. A chemical graph alignment algorithm was applied to detect the positions and numbers of chemical changes between two chemical compounds and exhibited better performance in distinguishing regioisomers [69].
Toward better prediction of reaction sequences in metabolic pathways
Reactions in metabolic pathways do not occur at random: reaction sequences exhibit many conserved patterns, termed reaction modules [70,71]. The concept of “enzymatic reaction-likeness” can be generalized as “k-step reaction sequence-likeness”, predicting how many (k) reactions are required to convert the starting compound into the goal compound [72]. Intermediate compounds are also predicted by a recursive procedure using step-specific classifiers (that predict the n-th compounds in the k-step reactions) based on chemical substructures. This resembles the compound-filling framework but is actually based on a reaction-filling framework because the intermediate compounds are taken from a pre-defined set of compounds. The advantage of k-step reaction sequence-likeness is the ability to predict the number of reaction steps between given pairs of compounds. Thus, combined with the compound-filling framework, this method would provide the computational efficiency necessary to consider unknown intermediates in the de novo metabolic pathway reconstruction.
Experimental annotation of metabolites using substrate-product relationships
The concept of substrate-product pairs has proven valuable to the experimental annotation of metabolites. Morreel et al. [73] assigned structures to the peaks derived from reversed-phase LC-negative electrospray ionization-MS profiling of plants by considering retention time and mass differences. This work demonstrated the potential of substrate-product pairs to experimentally identify metabolites and the enzymes responsible for reactions.
Prediction of EC numbers from substrate-product pairs
Having assigned a substrate-product relationship, corresponding enzyme genes may be predicted using two different strategies. If at least one enzyme is already known to catalyze the corresponding reaction, its amino acid sequence can be used to search for a homologous enzyme in the organism (genome) of interest (Fig. 2). Where no enzyme is known to catalyze the corresponding reaction, chemical structure information must be used to predict the corresponding enzyme (Fig. 5).
Figure 5.
Prediction of enzymes from chemical structures. Different from the reference-based metabolic pathway reconstruction, this process aims to predict enzymes where no enzymes have been identified so far. In order to achieve this goal, it is necessary to find known reactions (not proteins) that are similar to the putative reactions.
In general, similar reactions are present within each EC sub-subclass, designated by the third digit of the EC number (Fig. 1). The first method to automatically suggest a list of corresponding EC sub-subclasses was devised in 2004 and used a template-matching method of chemical transformation rules between known and query substrate-product pairs [74]. This prediction program, named E-zyme, was later improved in terms of both accuracy and coverage by applying multi-layered partial template matching and a weighted major voting scheme [75]. Note that this prediction of EC numbers is based not on amino acid sequence similarity but on chemical structure similarity. Several other methods to predict EC numbers from chemical structures have been subsequently developed [76–83], including EC-BLAST [83], which allows one to search for enzymatic reactions with EC numbers that are similar to the query reaction based on bond-change, reaction-center, or reaction-structure similarity.
Enzyme classification and enzyme protein similarity
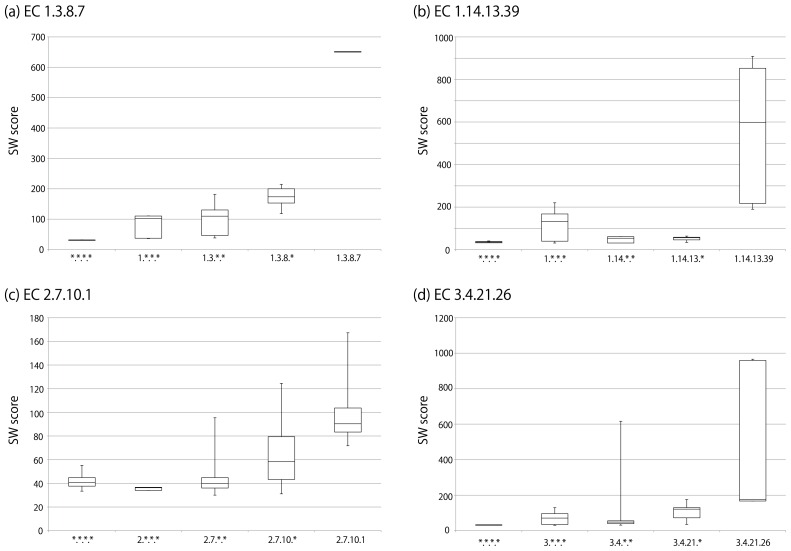
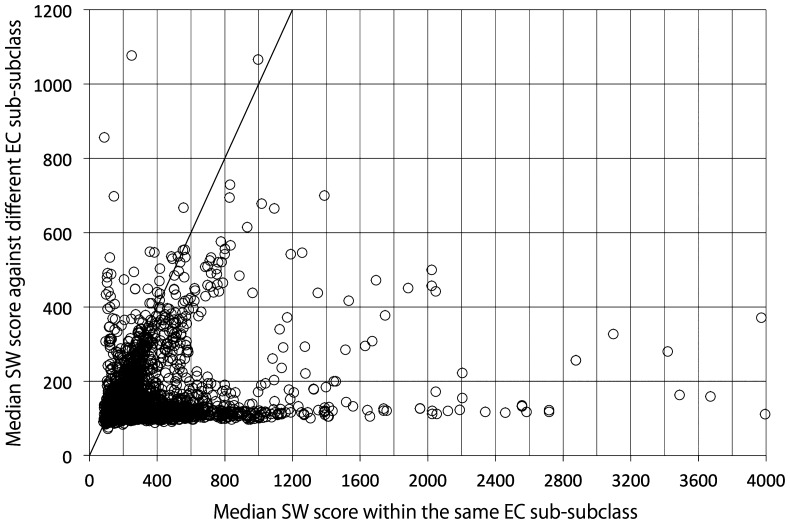
Enzyme classification by EC number is done according to a hierarchical structure (Fig. 1), and thus it is sometimes used as a measure of enzymatic reaction similarity. However, the purpose of the EC number system is to give unique names to enzymes, and therefore it is not appropriate to say that enzymes are dissimilar simply because their EC numbers are different. For example, as illustrated in Figure 6(a), the amino acid sequence of the enzyme labeled EC 1.3.8.7 is more similar to those of other EC 1.3.8 enzymes than to those of EC 1.3 enzymes belonging to other sub-subclasses. In such cases, it would be appropriate to use EC number as a proxy for enzyme similarity. However, there are many enzymes for which the EC hierarchy does not reflect enzyme similarity. For example, EC 1.14.13.39 enzymes are generally more similar to other oxidoreductases (EC 1 enzymes) than to other enzymes in EC 1.14 (Fig. 6(b)). Similarly, EC 2.7.10.1 enzymes are generally more similar to non-transferase enzymes than to other transferases (EC 2 enzymes) (Fig. 6(c)). EC 3.4.21.26 enzymes are generally less similar to other proteases (EC 3.4 enzymes) than to those in EC 3.4.21 but are exceptionally similar to some proteases that are not classified in EC 3.4.21 (Fig. 6(d)), consistent with the NC-IUBMB’s statement that EC 3.4 enzymes remain inadequately covered [20]. These examples are not rare cases, as considerable numbers of enzymes in different EC sub-subclasses are more similar to each other than they are to other enzymes in the same EC sub-subclasses (Fig. 7). This reflects the widely known fact that EC classification and protein 3D structures are not well-correlated [84].
Figure 6.
Smith-Waterman (SW) scores of enzyme proteins in different EC levels. The boxplot represents the minimum, the first quartile, the second quartile (median), the third quartile, and the maximum of SW scores derived from SSEARCH36 program with the default settings. (a) The “1.3.8.7” column shows the distribution of SW scores among the enzyme proteins that are given the annotation label “EC 1.3.8.7” in various organisms in KEGG. The “1.3.8.*” column shows the distribution of SW scores between the enzyme proteins with the label “EC 1.3.8.7” against the enzyme proteins with the “EC 1.3.8.*” label (belonging to the EC 1.3.8 sub-subclass but are not given EC 1.3.8.7) in various organisms. The “1.3.*.*” column shows the distribution of SW scores between the proteins of “EC 1.3.8.7” against the proteins with the “EC 1.3.*.*” label (belonging to the EC 1.3 subclass but not to EC 1.3.8 sub-subclass) in various organisms. The “1.*.*.*” column shows the distribution of SW scores between the proteins of “EC 1.3.8.7” against the proteins with the “EC 1.*.*.*”label (belonging to the EC 1 class but not to EC 1.3 subclass) in various organisms. The “*.*.*.*” column shows the distribution of SW scores between the proteins of “EC 1.3.8.7” against the enzymes that are not in EC 1 class in various organisms. The same procedures were conducted for (b) EC 1.14.13.39, (c) EC 2.7.10.1, and (d) EC 3.4.21.26.
Figure 7.
Comparison of SW scores between the enzyme proteins within the same EC sub-subclasses and in different EC sub-subclasses. Each circle represents a group of enzyme proteins having the same EC number in various organisms. The horizontal axis represents the median SW scores between proteins within the same EC sub-subclasses. The vertical axis represents the median SW scores against the proteins in different EC sub-subclasses. The diagonal line represents where the SW scores in horizontal and vertical axes are the same. SW scores were derived from SSEARCH36 program with the default settings.
Prediction of enzyme proteins from substrateproduct pairs
A number of methods have been proposed to assign EC sub-subclasses from chemical transformations, but they do not suggest catalytic enzyme gene/protein sequences. Recently, an extension of the E-zyme strategy was proposed to search for enzyme genes that catalyze the most similar reactions to those of the query substrate-product pair [85]. This method assesses enzyme specificity by scoring the substructures that are preserved in reactions catalyzed by orthologous enzymes, enabling prediction of enzyme protein orthologs from a query substrate-product pair.
Ontology to describe substrate-product pairs
Proteins, including enzymes, often consist of conserved partial structures, such as protein domains. Similarly, enzymatic reactions also consist of conserved partial reaction characteristics. For example, EC 5 (isomerases) includes intramolecular oxidoreductases (EC 5.3), intramolecular transferases (EC 5.4), and intramolecular lyases (EC 5.5), which share reaction characteristics with oxidoreductases (EC 1), transferases (EC 2), and lyases (EC 4), respectively. In order to organize the data according to these reaction characteristics, an ontology—the Enzymatic Reaction Ontology for Partial Information (PIERO) [86]—was recently developed. This ontology focuses on reducing reaction equations to substrate-product pairs and providing names to those pairs. Further improvement of this ontology would allow the substrate-product terminology to be used for building relationships between putative reactions that are not yet fully characterized and identification of corresponding enzyme proteins.
Concluding remarks
In this review, we briefly discussed the challenges involved in metabolic pathway reconstruction. Understanding what information is available is critical, as it determines potential strategies, i.e., reference-based or de novo. Additional research and development is still needed in order to predict enzymatic reactions from enzyme proteins and vice versa. Sequence homology and best-hit strategies are successful in many cases, but it must be remembered that a single amino acid mutation may alter enzyme activity and/or specificity. While some studies have attempted to analyze reaction similarities, methods remain insufficient for prediction of enzyme proteins from reactions alone. In order to understand broad patterns in metabolism, information regarding enzymes and reactions must be effectively organized and the structure-function relationships of enzyme proteins must be thoroughly and systematically analyzed.
Significance.
This review summarizes the in silico studies for reference-based and de novo metabolic pathway reconstruction problems. The availability of well-characterized enzymatic reactions determines which metabolic pathway reconstruction strategy we can choose to understand metabolic pathways in an organism of interest. The importance of the classification and structure-function relationships of enzymes is also discussed.
Acknowledgment
This work was supported by MEXT/JSPS Kakenhi (25108714) and the JST/MEXT Program to Promote the Tenure Track System in Tokyo Institute of Technology to M. K, and was supported in part by Japan Science and Technology Agency - Core Research for Evolutional Science and Technology (JST-CREST) “Establishment of core technology for the preservation and regeneration of marine biodiversity and ecosystems” to S. G.
Footnotes
Conflict of Interest
None declared.
Author Contributions
M. K. and S. G. collected the relevant articles for this review, and they drafted, completed and approved the manuscript.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klassen JL. Microbial secondary metabolites and their impacts on insect symbioses. Curr Opin Insect Sci. 2014;4:15–22. doi: 10.1016/j.cois.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Heidel-Fischer HM, Vogel H. Molecular mechanisms of insect adaptation to plant secondary compounds. Curr Opin Insect Sci. 2015;8:8–14. doi: 10.1016/j.cois.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Dixon RA, Strack D. Phytochemistry meets genome analysis, and beyond. Phytochemistry. 2003;62:815–816. doi: 10.1016/s0031-9422(02)00712-4. [DOI] [PubMed] [Google Scholar]
- 5.Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 6.Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, et al. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- 7.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald AG, Boyce S, Tipton KF. ExplorEnz: the primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009;37:D593–D597. doi: 10.1093/nar/gkn582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 11.Tolstikov VV, Fiehn O. Analysis of highly polar compounds of plant origin: combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal Biochem. 2002;301:298–307. doi: 10.1006/abio.2001.5513. [DOI] [PubMed] [Google Scholar]
- 12.von Roepenack-Lahaye E, Degenkolb T, Zerjeski M, Franz M, Roth U, Wessjohann L, et al. Profiling of Arabidopsis secondary metabolites by capillary liquid chromatography coupled to electrospray ionization quadrupole time-of-flight mass spectrometry. Plant Physiol. 2004;134:548–559. doi: 10.1104/pp.103.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 14.Sato S, Soga T, Nishioka T, Tomita M. Simultaneous determination of the main metabolites in rice leaves using capillary electrophoresis mass spectrometry and capillary electrophoresis diode array detection. Plant J. 2004;40:151–163. doi: 10.1111/j.1365-313X.2004.02187.x. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 16.Fernie AR. The future of metabolic phytochemistry: larger numbers of metabolites, higher resolution, greater understanding. Phytochemistry. 2007;68:2861–2880. doi: 10.1016/j.phytochem.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Nakabayashi R, Saito K. Metabolomics for unknown plant metabolites. Anal Bioanal Chem. 2013;405:5005–5011. doi: 10.1007/s00216-013-6869-2. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman-Ostenhof O. Suggestions for a more rational classification and nomenclature of enzymes. Adv Enzymol Relat Subj Biochem. 1953;14:219–220. doi: 10.1002/9780470122594.ch7. [DOI] [PubMed] [Google Scholar]
- 19.Dixon M, Webb EC. Enzymes. Longmans Green; London: Academic Press; New York: 1958. [Google Scholar]
- 20.McDonald AG, Tipton KF. Fifty-five years of enzyme classification: advances and difficulties. FEBS J. 2014;281:583–592. doi: 10.1111/febs.12530. [DOI] [PubMed] [Google Scholar]
- 21.Chang A, Schomburg I, Placzek S, Jeske L, Ulbrich M, Xiao M, et al. BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res. 2015;43:D439–446. doi: 10.1093/nar/gku1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgat A, Axelsen KB, Lombardot T, Alcántara R, Aimo L, Zerara M, et al. Updates in Rhea—a manually curated resource of biochemical reactions. Nucleic Acids Res. 2015;43:D459–464. doi: 10.1093/nar/gku961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotera M, Goto S, Kanehisa M. Predictive genomic and metabolomic analysis for the standardization of enzyme data. Perspectives in Science. 2014;1:24–32. [Google Scholar]
- 25.Bono H, Goto S, Fujibuchi W, Ogata H, Kanehisa M. Systematic Prediction of Orthologous Units of Genes in the Complete Genomes. Genome Inform Ser Workshop Genome Inform. 1998;9:32–40. [PubMed] [Google Scholar]
- 26.Galperin MY, Koonin EV. Functional genomics and enzyme evolution. Homologous and analogous enzymes encoded in microbial genomes. Genetica. 1999;106:159–170. doi: 10.1023/a:1003705601428. [DOI] [PubMed] [Google Scholar]
- 27.Dandekar T, Schuster S, Snel B, Huynen M, Bork P. Pathway alignment: application to the comparative analysis of glycolytic enzymes. Biochem J. 1999;343:115–124. [PMC free article] [PubMed] [Google Scholar]
- 28.Forst CV, Schulten K. Evolution of metabolisms: a new method for the comparison of metabolic pathways using genomics information. J Comput Biol. 1999;6:343–360. doi: 10.1089/106652799318319. [DOI] [PubMed] [Google Scholar]
- 29.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kharchenko P, Vitkup D, Church GM. Filling gaps in a metabolic network using expression information. Bioinformatics. 2004;20:i178–i185. doi: 10.1093/bioinformatics/bth930. [DOI] [PubMed] [Google Scholar]
- 31.Green ML, Karp PD. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamanishi Y, Mihara H, Osaki M, Muramatsu H, Esaki N, Sato T, et al. Prediction of missing enzyme genes in a bacterial metabolic network. Reconstruction of the lysine-degradation pathway of Pseudomonas aeruginosa. FEBS J. 2007;274:2262–2273. doi: 10.1111/j.1742-4658.2007.05763.x. [DOI] [PubMed] [Google Scholar]
- 33.Kotera M, Yamanishi Y, Moriya Y, Kanehisa M, Goto S. GENIES: gene network inference engine based on supervised analysis. Nucleic Acids Res. 2012;40:W162–W167. doi: 10.1093/nar/gks459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada T, Waller AS, Raes J, Zelezniak A, Perchat N, Perret A, et al. Prediction and identification of sequences coding for orphan enzymes using genomic and metagenomic neighbours. Mol Syst Biol. 2012;8:581. doi: 10.1038/msb.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 37.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takami H, Taniguchi T, Moriya Y, Kuwahara T, Kanehisa M, Goto S. Evaluation method for the potential functionome harbored in the genome and metagenome. BMC Genomics. 2012;13:699. doi: 10.1186/1471-2164-13-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Kim TY, Sohn SB, Kim YB, Kim WJ, Lee SY. Recent advances in reconstruction and applications of genome-scale metabolic models. Curr Opin Biotechnol. 2012;23:617–623. doi: 10.1016/j.copbio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Melchior F, Kindl H. Grapevine stilbene synthase cDNA only slightly differing from chalcone synthase cDNA is expressed in Escherichia coli into a catalytically active enzyme. FEBS Lett. 1990;268:17–20. doi: 10.1016/0014-5793(90)80961-h. [DOI] [PubMed] [Google Scholar]
- 43.Frank J, Holzwarth JF, Koch P, Vater J. Kinetics and molecular modelling of ligand binding to ribulose 1,5-bisphosphate carboxylase/oxygenase (RUBISCO) Berichte der Bunsengesellschaft für physikalische Chemie. 1996;100:2112–2116. [Google Scholar]
- 44.Oba Y, Ojika M, Inouye S. Firefly luciferase is a bifunctional enzyme: ATP-dependent monooxygenase and a long chain fatty acyl-CoA synthetase. FEBS Lett. 2003;540:251–254. doi: 10.1016/s0014-5793(03)00272-2. [DOI] [PubMed] [Google Scholar]
- 45.Overbeek R, Fonstein M, D’Souza M, Pusch G, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcotte E, Pellegrini M, Thompson M, Yeates T, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 47.Enright A, Iliopoulos I, Kyrpides N, Ouzounis C. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 48.Huynen M, Snel B, Lathe W, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osterman A, Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr Opin Chem Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 50.Kharchenko P, Vitkup D, Church G. Filling gaps in a metabolic network using expression information. Bioinformatics. 2004;20:i178–i185. doi: 10.1093/bioinformatics/bth930. [DOI] [PubMed] [Google Scholar]
- 51.Faulon JL, Sault AG. Stochastic generator of chemical structure. 3. Reaction network generation. J Chem Inf Comput Sci. 2001;41:894–908. doi: 10.1021/ci000029m. [DOI] [PubMed] [Google Scholar]
- 52.Satoh K, Funatsu K. A Novel Approach to Retrosynthetic Analysis Using Knowledge Bases Derived from Reaction Databases. J Chem Inf Comput Sci. 1999;39:316–325. [Google Scholar]
- 53.Darvas F. Predicting metabolic pathways by logic programming. J Mol Graph. 1988;6:80–86. [Google Scholar]
- 54.Talafous J, Sayre L, Mieyal J, Klopman G. A dictionary model of mammalian xenobiotic metabolism. J Chem Inf Comput Sci. 1994;34:1326–1333. doi: 10.1021/ci00022a015. [DOI] [PubMed] [Google Scholar]
- 55.Greene N, Judson P, Langowski J, Marchant C. Knowledge-based expert systems for toxicity and metabolism prediction: DEREK, StAR and METEOR. SAR QSAR Environ Res. 1999;10:299–314. doi: 10.1080/10629369908039182. [DOI] [PubMed] [Google Scholar]
- 56.Moriya Y, Shigemizu D, Hattori M, Tokimatsu T, Kotera M, Goto S, et al. PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 2010;38:W138–W143. doi: 10.1093/nar/gkq318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J, Ellis L, Wackett L. TheUniversity of Minnesota Pathway Prediction System: multi-level prediction and visualization. Nucleic Acids Res. 2011;39:W406–W411. doi: 10.1093/nar/gkr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fenner K, Gao J, Kramer S, Ellis L, Wackett L. Data-driven extraction of relative reasoning rules to limit combinatorial explosion in biodegradation pathway prediction. Bioinformatics. 2008;24:2079–2085. doi: 10.1093/bioinformatics/btn378. [DOI] [PubMed] [Google Scholar]
- 59.Oh M, Yamada T, Hattori M, Goto S, Kanehisa M. Systematic analysis of enzyme-catalyzed reaction patterns and prediction of microbial biodegradation pathways. J Chem Inf Model. 2007;47:1702–1712. doi: 10.1021/ci700006f. [DOI] [PubMed] [Google Scholar]
- 60.Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, et al. Nontargeted metabolome analysis by use of Fourier Transform Ion Cyclotron Mass Spectrometry. OMICS. 2002;6:217–234. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- 61.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hatzimanikatis V, Li C, Ionita J, Henry C, Jankowski M, Broadbelt L. Exploring the diversity of complex metabolic networks. Bioinformatics. 2005;21:1603–1609. doi: 10.1093/bioinformatics/bti213. [DOI] [PubMed] [Google Scholar]
- 63.Nakamura M, Hachiya T, Saito Y, Sato K, Sakakibara Y. An efficient algorithm for de novo predictions of biochemical pathways between chemical compounds. BMC Bioinformatics. 2012;13:S8. doi: 10.1186/1471-2105-13-S17-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kotera M, McDonald A, Boyce S, Tipton K. Eliciting possible reaction equations and metabolic pathways involving orphan metabolites. J Chem Inf Model. 2008;48:2335–2349. doi: 10.1021/ci800213g. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka K, Nakamura K, Saito T, Osada H, Hirai A, Takahashi H, et al. Metabolic pathway prediction based on inclusive relation between cyclic substructures. Plant Biotechnol. 2009;26:459–468. [Google Scholar]
- 66.Kotera M, Tabei Y, Yamanishi Y, Tokimatsu T, Goto S. Supervised de novo reconstruction of metabolic pathways from metabolome-scale compound sets. Bioinformatics. 2013;29:i135–i144. doi: 10.1093/bioinformatics/btt244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beisken S, Meinl T, Wiswedel B, de Figueiredo LF, Berthold M, Steinbeck C. KNIME-CDK: Workflow-driven cheminformatics. BMC Bioinformatics. 2013;14:257. doi: 10.1186/1471-2105-14-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kotera M, Tabei Y, Yamanishi Y, Moriya Y, Tokimatsu T, Kanehisa M, et al. KCF-S: KEGG Chemical Function and Substructure for improved interpretability and prediction in chemical bioinformatics. BMC Syst Biol. 2013;7:S2. doi: 10.1186/1752-0509-7-S6-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kotera M, Tabei Y, Yamanishi Y, Muto A, Moriya Y, Tokimatsu T, et al. Metabolome-scale prediction of intermediate compounds in multistep metabolic pathways with a recursive supervised approach. Bioinformatics. 2014;30:i165–i174. doi: 10.1093/bioinformatics/btu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muto A, Kotera M, Tokimatsu T, Nakagawa Z, Goto S, Kanehisa M. Modular architecture of metabolic pathways revealed by conserved sequences of reactions. J Chem Inf Model. 2013;53:613–622. doi: 10.1021/ci3005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sorokina M, Medigue C, Vallenet D. A new network representation of the metabolism to detect chemical transformation modules. BMC Bioinformatics. 2015;16:385. doi: 10.1186/s12859-015-0809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamanishi Y, Tabei Y, Kotera M. Metabolome-scale de novo pathway reconstruction using regioisomer-sensitive graph alignments. Bioinformatics. 2015;31:i161–i170. doi: 10.1093/bioinformatics/btv224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morreel K, Saeys Y, Dima O, Lu F, Van de Peer Y, Vanholme R, et al. Systematic structural characterization of metabolites in Arabidopsis via candidate substrate-product pair networks. Plant Cell. 2014;26:929–945. doi: 10.1105/tpc.113.122242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kotera M, Okuno Y, Hattori M, Goto S, Kanehisa M. Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J Am Chem Soc. 2004;126:16487–16498. doi: 10.1021/ja0466457. [DOI] [PubMed] [Google Scholar]
- 75.Yamanishi Y, Hattori M, Kotera M, Goto S, Kanehisa M. E-zyme: predicting potential EC numbers from the chemical transformation pattern of substrate-product pairs. Bioinformatics. 2009;25:i179–i186. doi: 10.1093/bioinformatics/btp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Boyle NM, Holliday GL, Almonacid DE, Mitchell JB. Using reaction mechanism to measure enzyme similarity. J Mol Biol. 2007;368:1484–1499. doi: 10.1016/j.jmb.2007.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Latino DA, Aires-de-Sousa J. Assignment of EC numbers to enzymatic reactions with MOLMAP reaction descriptors and random forests. J Chem Inf Model. 2009;49:1839–1846. doi: 10.1021/ci900104b. [DOI] [PubMed] [Google Scholar]
- 78.Egelhofer V, Schomburg I, Schomburg D. Automatic assignment of EC numbers. PLoS Comput Biol. 2010;6:e1000661. doi: 10.1371/journal.pcbi.1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu X, Yan A, Tan T, Sacher O, Gasteiger J. Similarity perception of reactions catalyzed by oxidoreductases and hydrolases using different classification methods. J Chem Inf Model. 2010;50:1089–1100. doi: 10.1021/ci9004833. [DOI] [PubMed] [Google Scholar]
- 80.Hu QN, Zhu H, Li X, Zhang M, Deng Z, Yang X, et al. Assignment of EC numbers to enzymatic reactions with reaction difference fingerprints. PLoS ONE. 2012;7:e52901. doi: 10.1371/journal.pone.0052901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nath N, Mitchell JB. Is EC class predictable from reaction mechanism? BMC Bioinformatics. 2012;13:60. doi: 10.1186/1471-2105-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matsuta Y, Ito M, Tohsato Y. ECOH: an enzyme commission number predictor using mutual information and a support vector machine. Bioinformatics. 2013;29:365–372. doi: 10.1093/bioinformatics/bts700. [DOI] [PubMed] [Google Scholar]
- 83.Rahman SA, Cuesta SM, Furnham N, Holliday GL, Thornton JM. EC-BLAST: a tool to automatically search and compare enzyme reactions. Nat Methods. 2014;11:171–174. doi: 10.1038/nmeth.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.George RA, Spriggs RV, Thornton JM, Al-Lazikani B, Swindells MB. SCOPEC: a database of protein catalytic domains. Bioinformatics. 2004;20:i130–i136. doi: 10.1093/bioinformatics/bth948. [DOI] [PubMed] [Google Scholar]
- 85.Moriya Y, Yamada T, Okuda S, Nakagawa Z, Kotera M, Tokimatsu T, et al. Identification of Enzyme Genes Using Chemical Structure Alignments of Substrate-Product Pairs. J Chem Inf Model. 2016;56:510–516. doi: 10.1021/acs.jcim.5b00216. [DOI] [PubMed] [Google Scholar]
- 86.Kotera M, Nishimura Y, Nakagawa Z, Muto A, Moriya Y, Okamoto S, et al. PIERO ontology for analysis of biochemical transformations: effective implementation of reaction information in the IUBMB enzyme list. J Bioinform Comput Biol. 2014;12:1442001. doi: 10.1142/S0219720014420013. [DOI] [PubMed] [Google Scholar]