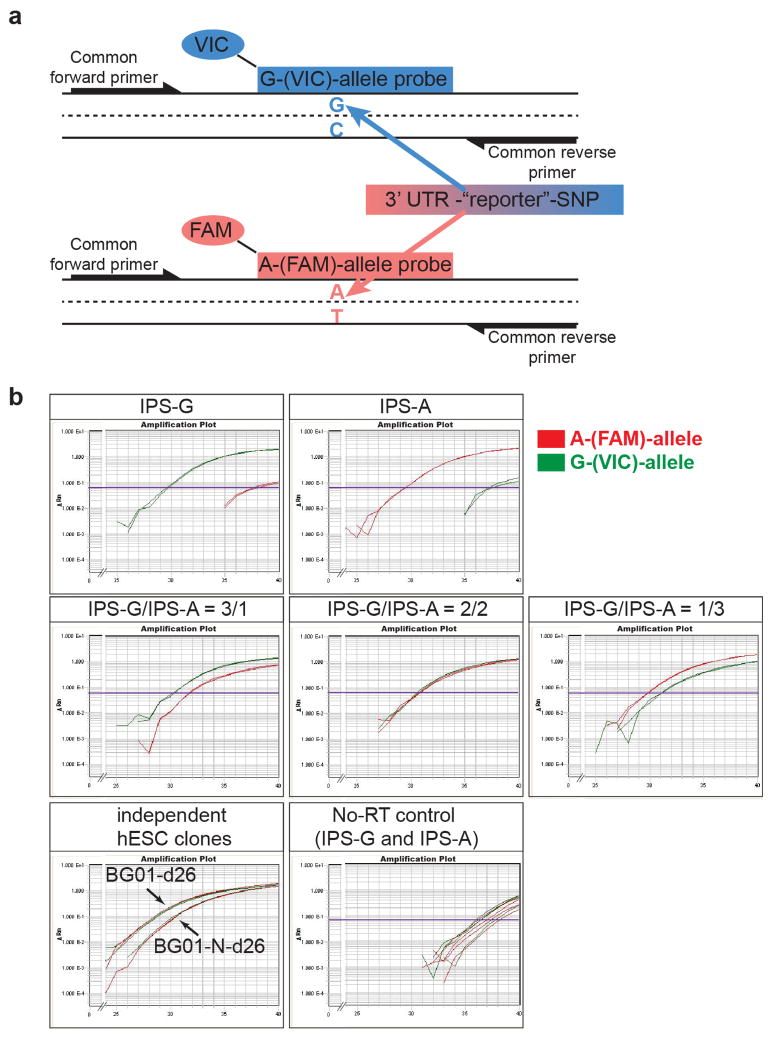
Extended data Fig. 1. Analysis of allele-specific expression of SNCA using quantitative reverse transcription polymerase chain reaction (qRT-PCR).
(a) Schematic illustration of the quantitative allele-specific SNCA expression analysis using a common primer pair and allele-specific Taqman® probes conjugated with different fluorophores to specifically detect a reporter-SNP (rs356165) in the 3′UTR of SNCA in a multiplex reaction. As indicated, 6-carboxyfluorescein (FAM) and 4,7,2′-trichloro-7′-phenyl-6-carboxyfluorescein (VIC) were used to detect the A- and G-allele respectively. (b) Representative multiplex qRT-PCR reactions (in duplicates) measuring allele-specific SNCA expression of allele-biased samples described in Fig. 1c,d. Allele-biased samples were generated by mixing hIPSC-derived neurons homozygous for either the A-(IPS-A) or G-allele (IPS-G) at rs356165 at indicated ratios. Also included is a plot showing c-DNAs synthesized in the absence of SuperScript® reverse transcriptase (no-RT) to control for genomic DNA contaminations. Plots are displayed as reporter dye fluorescence signal (ΔRn) in log scale as a function of run cycle.