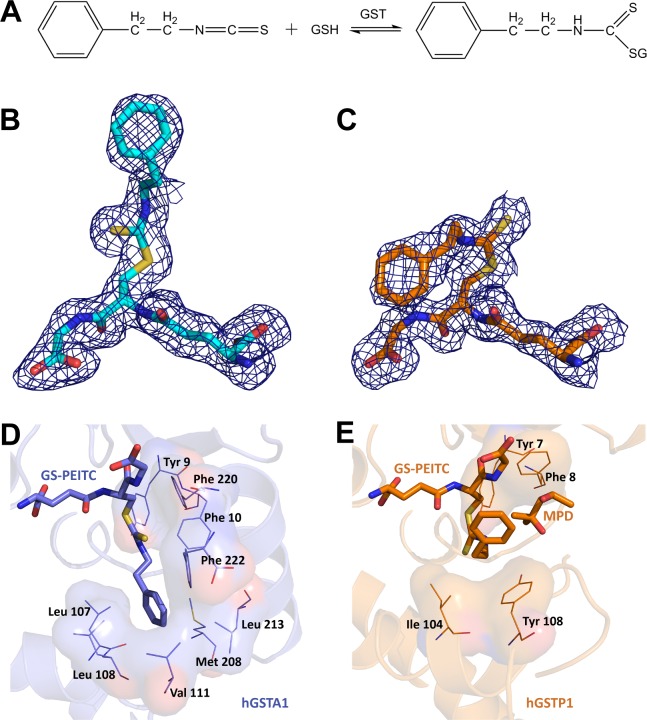
Fig 1. Distinct Binding Modes of GS-PEITC in GST Enzymes.
(A) GST catalyzes the conjugation of PEITC with GSH in both forward (formation of GS-PEITC) and reverse (dissociation of GS-PEITC) directions. (B,C) The GS-PEITC adduct bound in the active site of hGSTA1 and hGSTP1, respectively, is illustrated as a stick model in atomic color scheme (nitrogen in blue, carbon in cyan (hGSTA1) or orange (hGSTP1), oxygen in red, and sulfur in yellow) outlined with annealed omit map (blue mesh, Fo-Fc electron density contoured at 2.0 σ). Note the distinct conformations of GS-PEITC in the two GST active sites. (D) In the hGSTA1:GS-PEITC structure (PDB entry 5JCU), the phenethyl moiety of GS-PEITC docks into a hydrophobic pocket formed by the side chains of Leu107, Leu108, Val111, Met208, Leu213, Phe222, Phe10 and Phe220. (E) In the hGSTP1:GS-PEITC structure (PDB entry 5JCW), however, it points toward the solvent channel, stacking with the hydrophobic side chains of Ile104 and Tyr108 and in contact with a 2-methyl-2,4-pentanediol (MPD) molecule from cryoprotectant. In panels D and E, ligand molecules are illustrated as thick sticks, amino acid residues as thin sticks outlined with their surfaces, and protein structures as ribbon diagrams in the background. Nitrogen atoms are shown in blue, oxygen in red, and sulfur in yellow. Carbon atoms are shown in blue in hGSTA1:GS-PEITC, but in orange in hGSTP1:GS-PEITC.