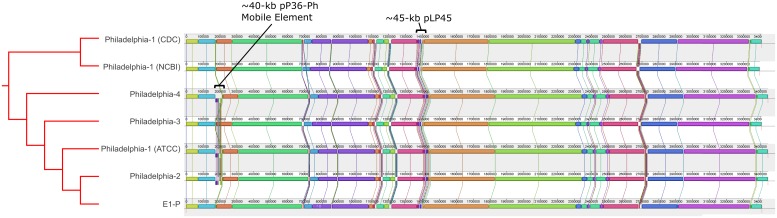
Fig 4. Mauve whole-genome alignment of L. pneumophila strains within the Philadelphia clade.
ProgressiveMauve was used to compare the fully assembled sequences of the Philadelphia historical Legionella strains as well as isolate E1-P. The minimum weight for pairwise LCBs (locally collinear blocks), which share common colors across genomes, was set to 100, otherwise, the program was run using default parameters as described in the Methods. The general clade organization, as well as the identity and location of the ~40-kb pP36-Ph and the ~45-kb pLP45 elements are shown. The general, expanded Philadelphia clade organization from Fig 1 is shown, therefore the phylogenetic distances are not to scale.