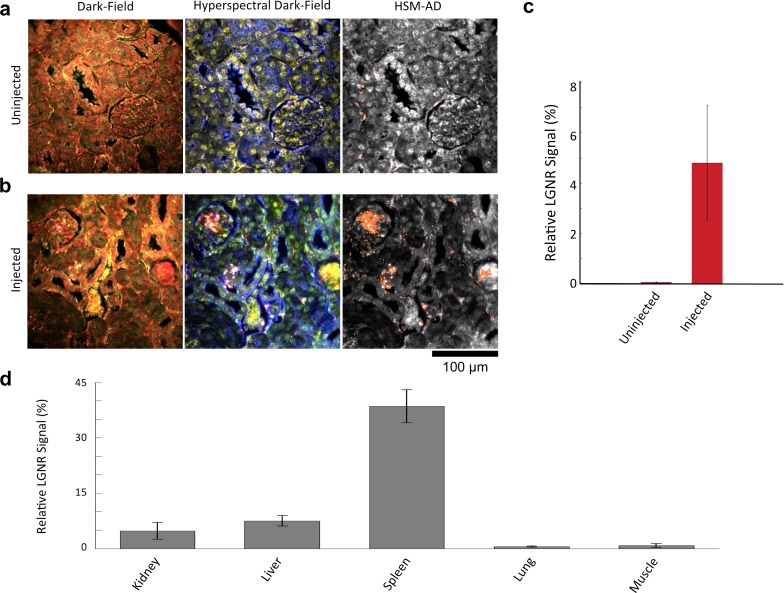
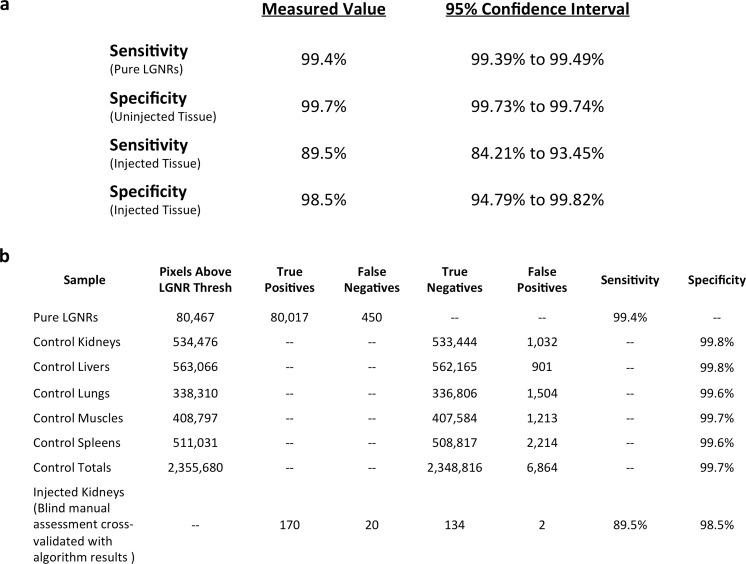
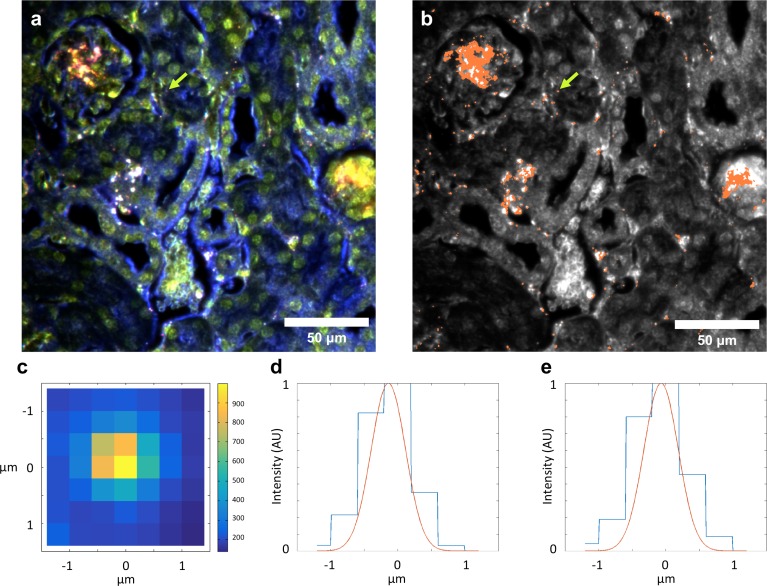
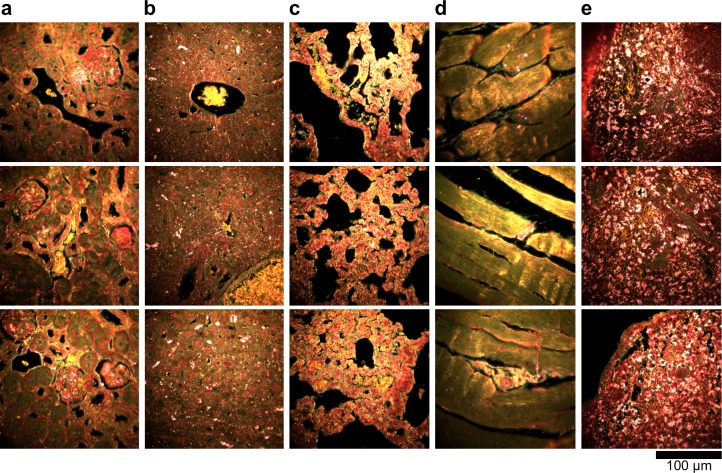
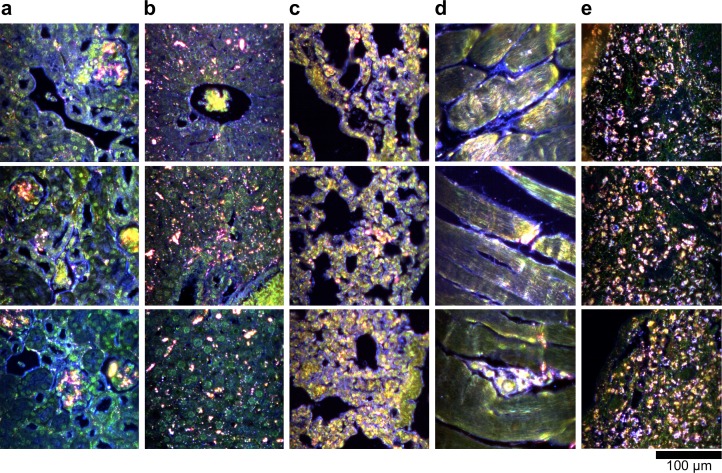
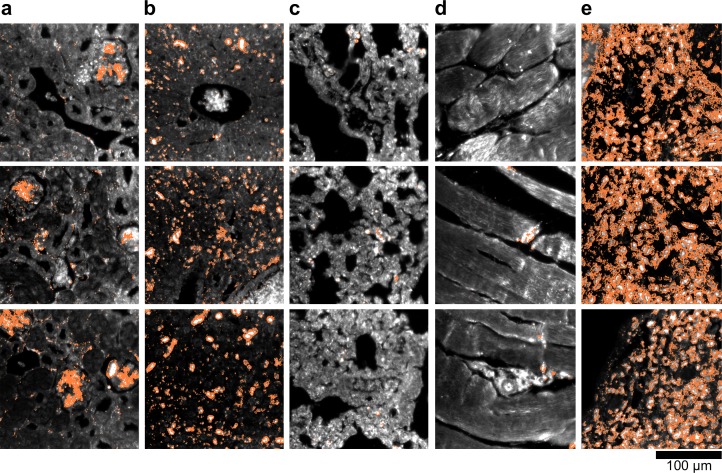
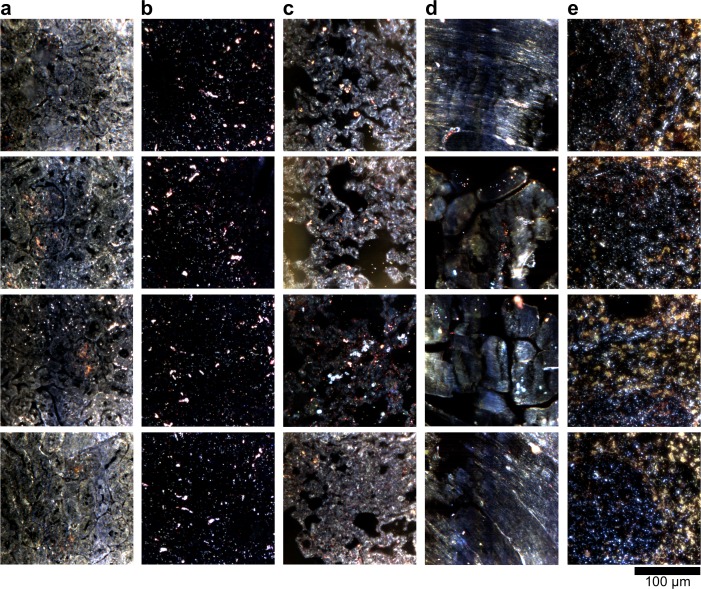
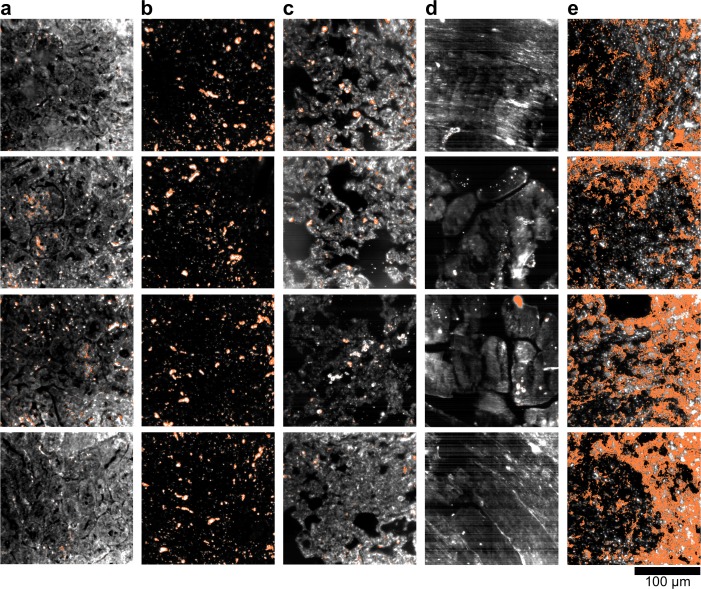
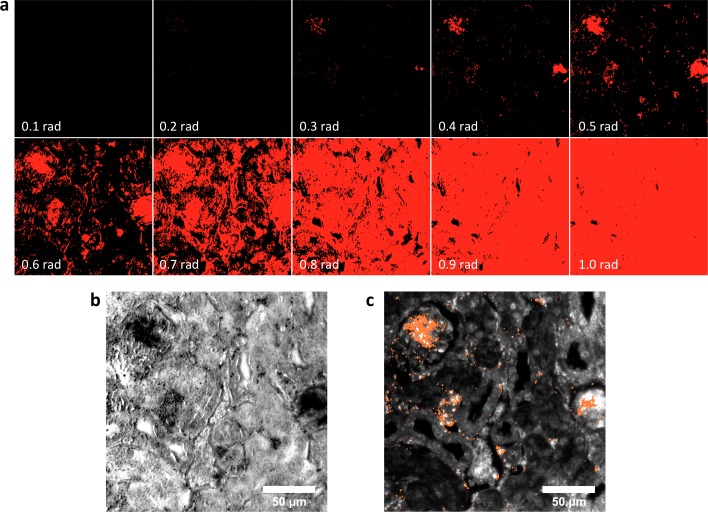
Figure 2. Sensitivity and specificity validation of HSM-AD.
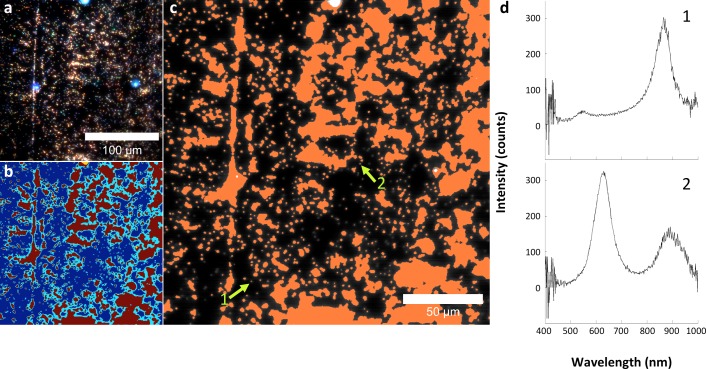
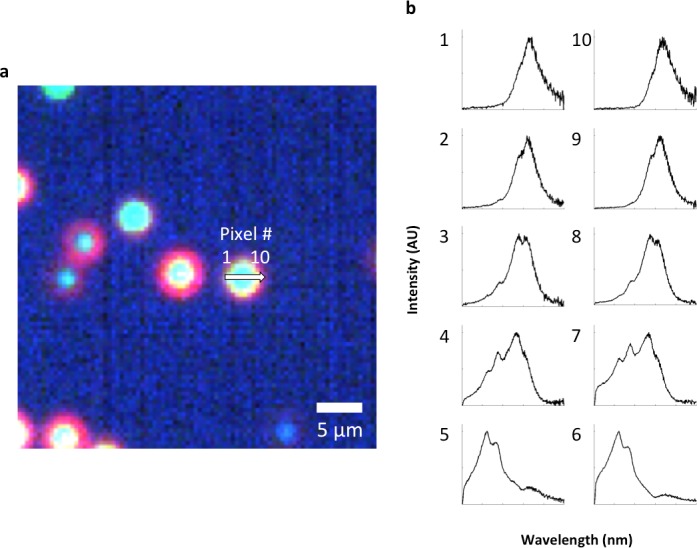
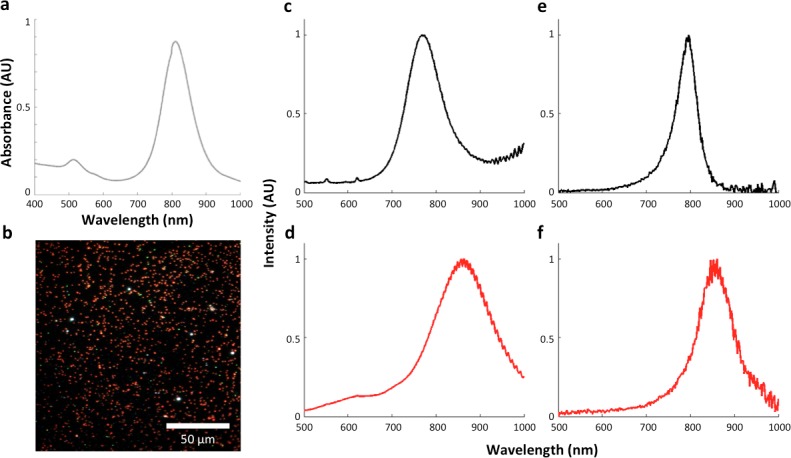
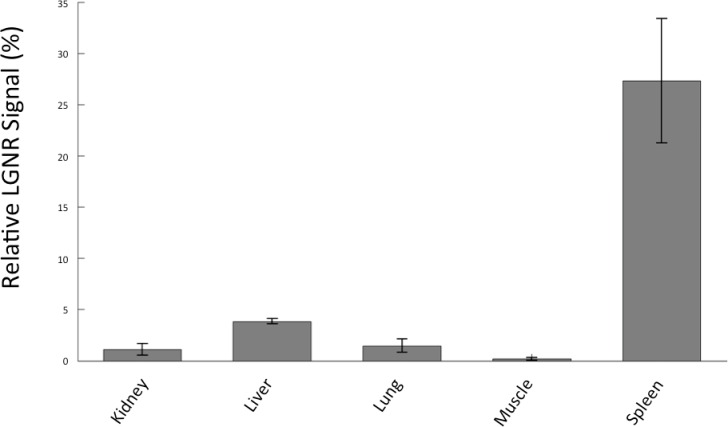
(a,b) Hematoxylin and Eosin (H&E) stained tissue samples (kidney) from uninjected (a) and injected (b) mice were imaged using dark-field and hyperspectral microscopy at 40x magnification and analyzed with HSM-AD to measure LGNR detection specificity and sensitivity. Conventional dark-field images highlight features including nuclei (salmon-pink), cytoplasm (green-brown), and erythrocytes (yellow-orange) within the tissues, but they reveal little information regarding the presence or absence of LGNRs. By comparison, putative LGNRs can be roughly identified as red-orange pixels in false-colored hyperspectral images, while nuclei and cytoplasm appear in green and indigo, respectively. (c) HSM-AD analysis of hyperspectral images demonstrates the absence of LGNRs in uninjected tissues and LGNR presence in injected samples (two-tailed Student’s t-test, p=0.054). Quantification of the relative LGNR signal from n = 4 tissue slices (representing a total of 1.04 million pixels) indicates that the false positive rate for LGNR detection (determined from uninjected tissues) is minimal. A detection specificity of 99.7% was determined from uninjected tissue sections, and a detection sensitivity of 99.4% was measured from samples of pure LGNRs analyzed using HSM-AD (Figure 2—figure supplement 1). (d) HSM-AD analysis of whole tissue sections (n = 4 for each tissue type) reveals quantitative differences in bulk LGNR uptake among various organs, in a manner analogous to conventional biodistribution methods. Quantitative data are presented as mean ± standard error of the mean (s.e.m.).
DOI: http://dx.doi.org/10.7554/eLife.16352.008