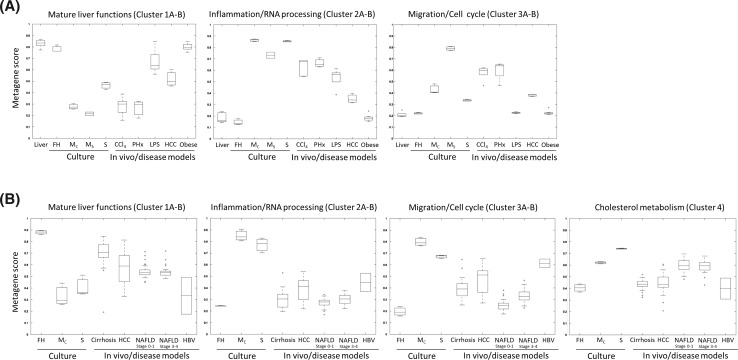
Fig. 6.
Quantitative analysis of the metagenes calculated for the three main clusters ‘mature liver functions,’ ‘inflammation/RNA processing’ and ‘migration/cell cycle’ in cultured hepatocytes and liver disease models in mouse (a) and human (b). Genes assigned to the metagenes ‘mature liver functions,’ ‘inflammation/RNA processing’ and ‘migration/cell cycle’ (and ‘cholesterol metabolism’ for human) were determined by selecting the top deregulated genes from each Fuzzy cluster group (Fig. 5; see ‘metagene analysis’ in ‘supplemental materials and methods’). The graphs indicate the metagene score for each condition, which corresponds to the mean scaled value of all genes of the corresponding cluster groups as identified in Fig. 5. For mouse (a), the analysis was performed based on expression levels in hepatocytes at day 1 in M C, M S and S, and the in vivo models of CCl4 intoxication at day 1 (CCl4), partial hepatectomy at day 1 (PHx), LPS-induced inflammation (LPS), hepatocellular carcinoma (HCC) and fatty liver (obese). For human (b), the analysis includes gene expression levels at day 1 in cultured hepatocytes, and on the in vivo disease models of cirrhosis, HCC, non-alcoholic fatty liver disease (NAFLD) on stages 0–1 and 2–4, and in HBV-infected liver tissue