Figure 1.
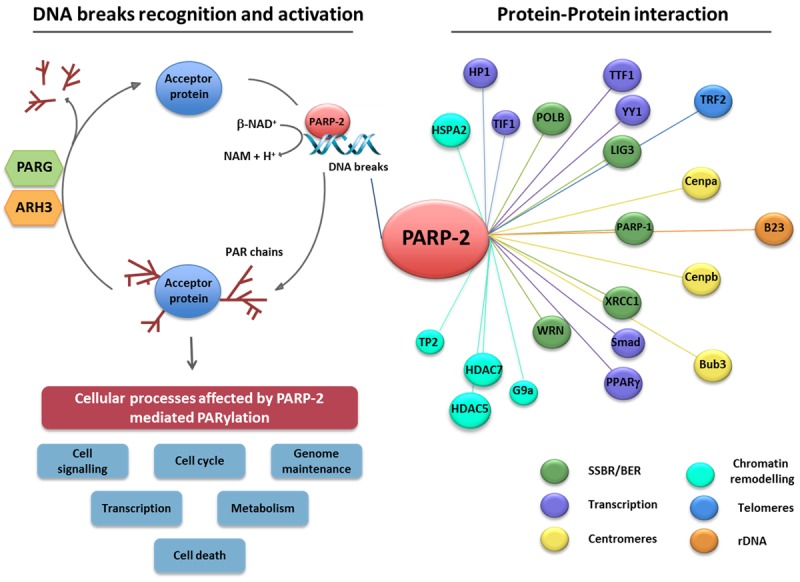
Poly(ADP-ribose) polymerase-2 (PARP-2) executes its biological functions via physical interaction with or PARylation of target proteins. PARP-2 is rapidly recruited to sites of DNA breakage leading to its enzymatic activation (left panel). Activated PARP-2, as well as other bona fide PARPs, hydrolyse NAD+ to nicotinamide (Nam) and one proton (H+), in doing so transferring an ADP-ribose moiety onto specific residues of protein substrates. The subsequent homopolymerisation of ADP-ribose molecules forms variable, long and branched chains of ADP-ribose polymer (PAR). Acceptor proteins subject to PARP-2-dependent PARylation are implicated in a wide range of biological processes, including genome maintenance, transcription, cell cycle regulation, cell death, cell signalling, and metabolism. The reverse reaction involves the rapid degradation of the PAR polymer by poly(ADP-ribose) glycohydrolase (PARG) and poly(ADP-ribose) hydrolase-3 (ARH3) which hydrolyse poly(ADP-ribose) into ADP-ribose units. Biochemical approaches and proteomics show that PARP-2 also physically associates with various protein partners (right panel) involved in different biological processes [9-19].
