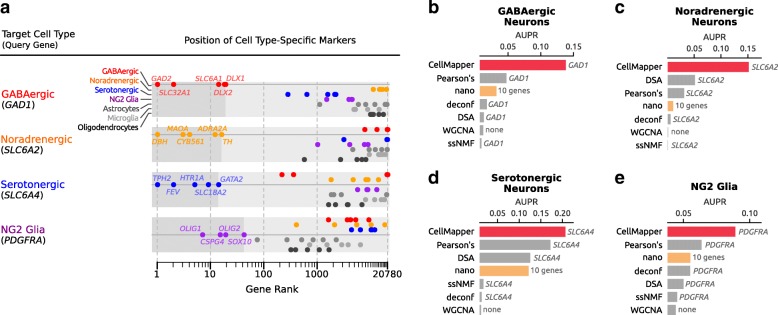
Fig. 2.
Application of CellMapper to brain cell types that are difficult to address by other methods. a CellMapper was applied to the Allen Brain Atlas dataset using the indicated query genes for four brain cell types. Dot charts display the rank of literature-defined cell-specific markers (positive controls) within CellMapper’s predictions for each cell type. Dots are colored based on their known primary cell type of expression. Dark gray shading covers the area (rank list) required to identify all positive control genes for each cell type. A similar analysis using query genes other than GAD1, SLC6A2, SLC6A4, and PDGFRA for the four cell types is provided in Additional file 16. b–e Performance evaluation of CellMapper and other computational methods to recover genes expressed in the four brain cell types. Each method was evaluated based on the recovery of an experimentally-defined [3–6] set of cell type-enriched genes in mouse, as quantified by the area under the precision recall curve (AUPR). WGCNA returns several modules of gene co-expression, the best performing WGCNA module is plotted for each cell type