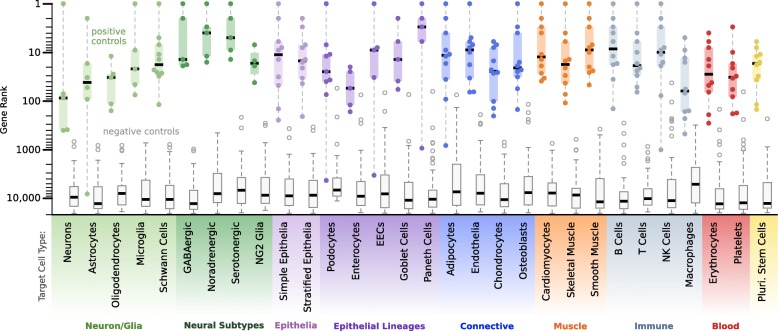
Fig. 3.
CellMapper is accurate across diverse cell types. CellMapper was applied using query genes for 30 cell types (Additional file 12); Tukey boxplots display the rank of 4–10 literature curated markers (positive controls; Additional file 13) and ≥48 negative control genes (Additional file 13 and housekeeping genes from [30]) for each cell type, demonstrating that CellMapper sensitively identified established cell type markers in every case. Filled circles represent the rank of all positive control genes; open gray circles represent negative control genes that fall outside 1.5 times the interquartile range of the other negative control genes (“outliers”). In only eight instances (0.5 %) was a negative control gene identified within the top 100 predictions for a cell type. EECs enteroendocrine cells