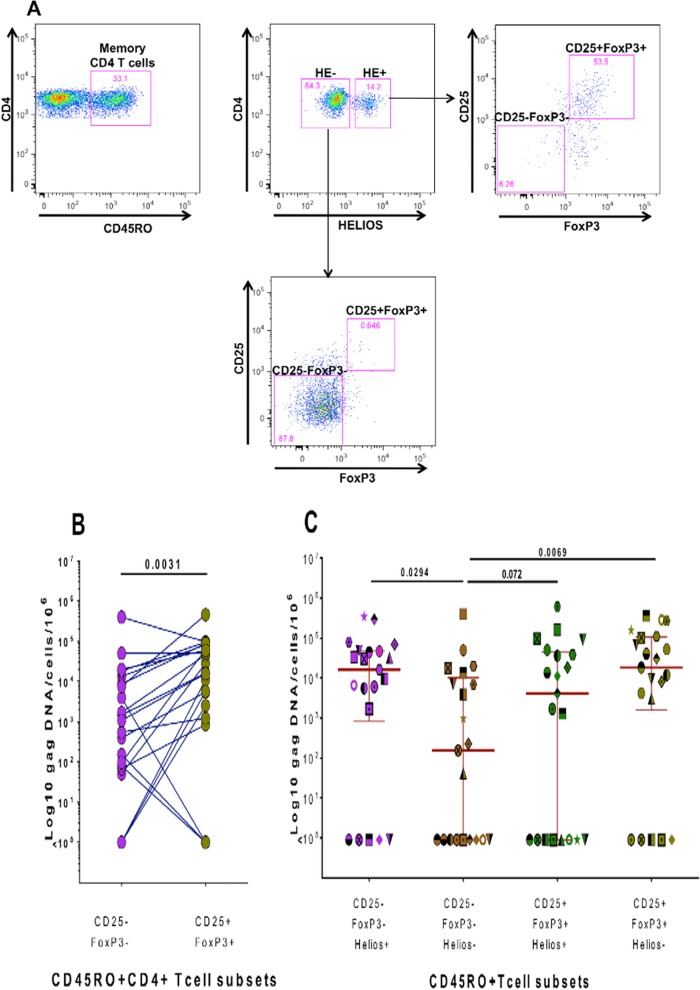
FIG 4.
Quantification of cell-associated HIV gag DNA in sorted memory CD4 T cell subsets. (A) Gating/sorting strategy used to sort different memory CD4 T cell populations delineated by Helios, CD25, and FoxP3 expression. (B) Numbers of gag DNA copies/106cells detected in CD25− FoxP3− and CD25+ FoxP3+ memory CD4 T cells from 21 different subjects. (C) Numbers of gag DNA copies/106cells detected in these memory CD4 T cell subsets further delineated by Helios expression. gag DNA within different CD4 T cell populations of the same subject was quantified during the same real-time (RT)-PCR run. Cryopreserved PBMCs from the WHIS and HISIS cohorts were used for cell sorting. Statistical analysis was performed using the Wilcoxon rank matched-pairs test.