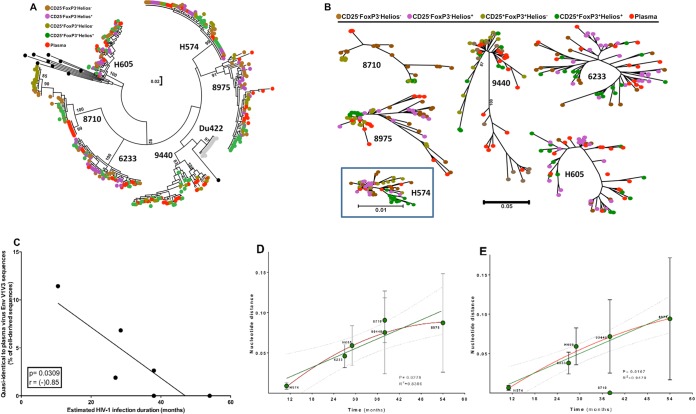
FIG 5.
Phylogenetic relationship of HIV envelope sequences derived from plasma and sorted memory CD4 T cell populations. Plasma- and cell-derived sequences of the highly variable EnvV1V3 region (Hxb 6559 to 7320) were amplified, cloned, sequenced (n = 384, Sanger method), and analyzed for 6 viremic subjects from the WHIS and HISIS cohorts with differing HIV infection durations (H574, H605, 6233K12 [6233], 9440A11 [9440], 8710U11 [8710], 8975T11 [8975]). (A and B) The phylogenetic relationship was inferred by the maximum-likelihood method based on the general time-reversible substitution model (GTR+G). (C) Correlation between frequency of cell-derived sequences that were quasi-identical to plasma-derived sequences and the estimated infection duration. (D and E) Linear regression analysis (green line) between the distance of the EnvV1V3 sequences derived from plasma to the sequences extracted from the corresponding cellular fractions and the estimated duration of infection (D) and plasma sequence diversity plotted against the estimated duration of infection (E). The red line indicates a nonlinear analysis performed using a second-order polynomial equation taking into account the best-fit values. The evolutionary distances were computed using the Kimura 2-parameter method (77) and are given in units of the number of base substitutions per site, including both transitions and transversions. The rate variation among sites was modeled with a gamma distribution. The analysis was conducted in MEGA6 (44). No sequence diversity was observed in the subject 8710 plasma fraction, probably because the number of viruses sampled in each PCR was very low (Table 2). We therefore excluded the results for subject 8710 from the linear regression analysis. P and r values were calculated with the Pearson two-tailed statistical test.