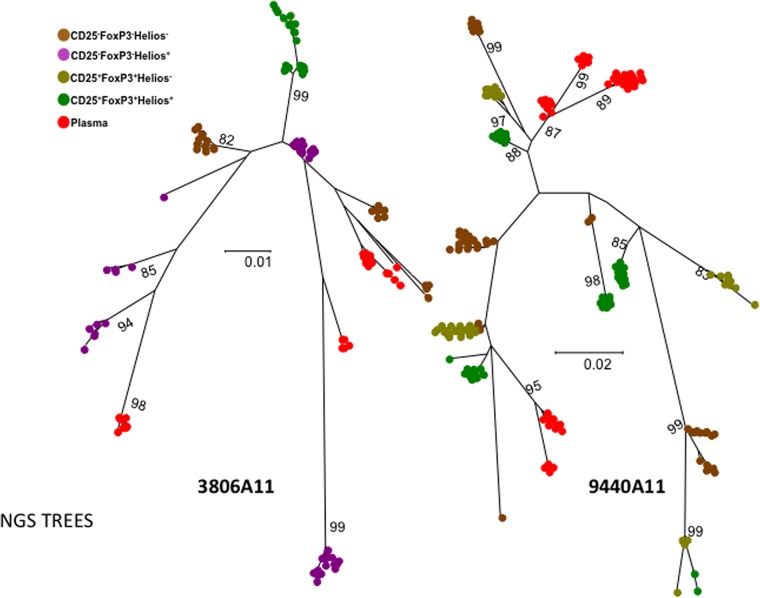
FIG 6.
Phylogenetic analyses of HIV envelope sequences derived from plasma and sorted memory CD4 T cell populations using next-generation sequencing. Shown are the phylogenetic analyses of EnvV1V3 sequences from the 50 most frequently detected sequences derived from either plasma or the different sorted memory CD4 T cell subsets for two viremic subjects of the WHIS cohort. The phylogenetic relationship was inferred by the maximum-likelihood method based on the general time-reversible substitution model (GTR+G). EnvV1V3 amplicons were directly subjected to next-generation sequencing. Quasispecies reconstruction was performed using the software QuasiRecomb. The methods used are described in detail in Materials and Methods.