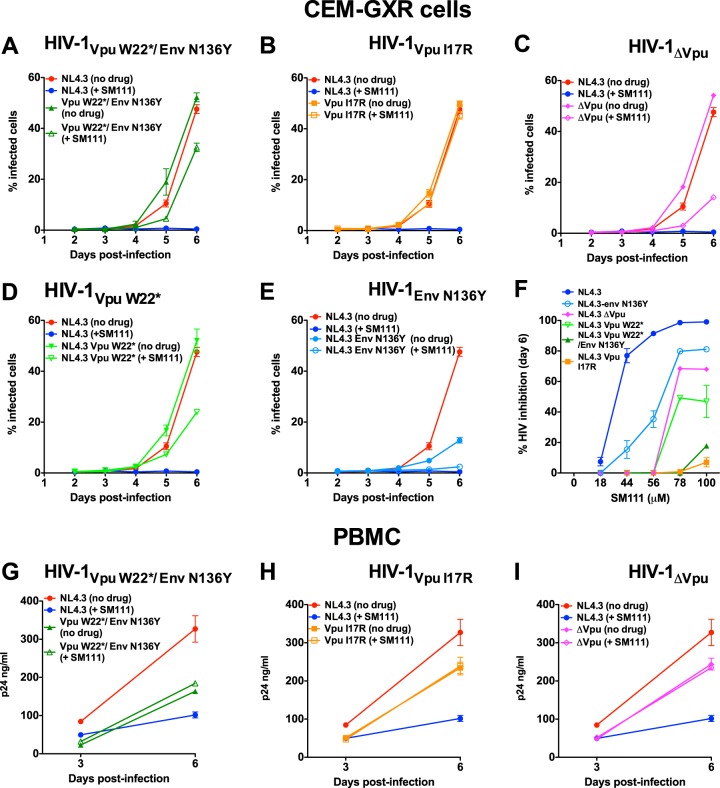
FIG 6.
Validation of SM111 resistance mutations in CEM-GXR cells and PBMC. (A to F) Replication of HIV-1VpuW22*/EnvN136Y (A), HIV-1VpuI17R (B), HIV-1ΔVpu (C), HIV-1VpuW22* (D), and HIV-1EnvN136Y (E) in CEM-GXR cells in comparison to HIV-1NL4.3 in the absence and presence of 100 μM SM111. (F) Percent inhibition of each virus strain at day 6 postinfection at various SM111 concentrations. (G to I) Replication of HIV-1VpuW22*/EnvN136Y (G), HIV-1VpuI17R (H), and HIV-1ΔVpu (I) in PBMC in comparison to HIV-1NL4.3 in the absence and presence of 56 μM SM111. Data are representative of three independent experiments performed in triplicate in CEM-GXR cells (A to F) or using PBMC from three different donors in triplicate (G to I). Data for HIV-1NL4.3 are shown repetitively to facilitate comparisons with replication of mutant strains. Error bars represent means ± SD.