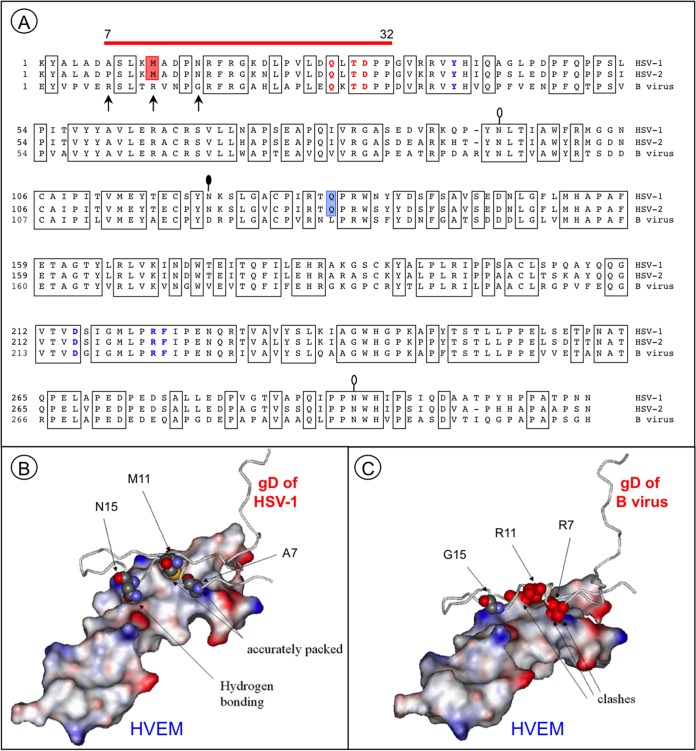
FIG 4.
Essential differences in protein-protein interfaces involving HSV-1 and B virus gD proteins and HVEM. (A) Alignment of the gD ectodomain residues of HSV-1 (KOS), HSV-2 (HG52), and the B virus lab strain (E2490). Conserved amino acid residues are boxed. The red line above the gD sequences indicates HVEM contact residues. The residues essential for HVEM-mediated and nectin-1-mediated HSV-1 entry are shown in bold red and bold blue, respectively. Red shading indicates the M11 residue, which is involved in the interaction with the HVEM “hot spot” residue Y28. Blue shading indicates the only residue in the IgV-core, Q132, that directly interacts with the nectin-1 C″ strand. The arrows point to the residues that most probably distort the HVEM-B virus gD interface. Lollipops mark the predicted N-glycosylation sites. The black-filled lollipop marks an N-glycosylation site that is absent in the B virus lab strain. (B) Resolved structure of the HSV-1 gD and HVEM complex (PDB code 1JMA). Residues 1 to 50 of gD are shown. The residues A7, M11, and N15 (shown in space-filling format) are accurately packed at the molecular surface of the receptor. (C) Model of the HVEM and B virus gD complex. Severe steric clashes of R7 and R11 are predicted, with no interactions between G15 and the receptor.