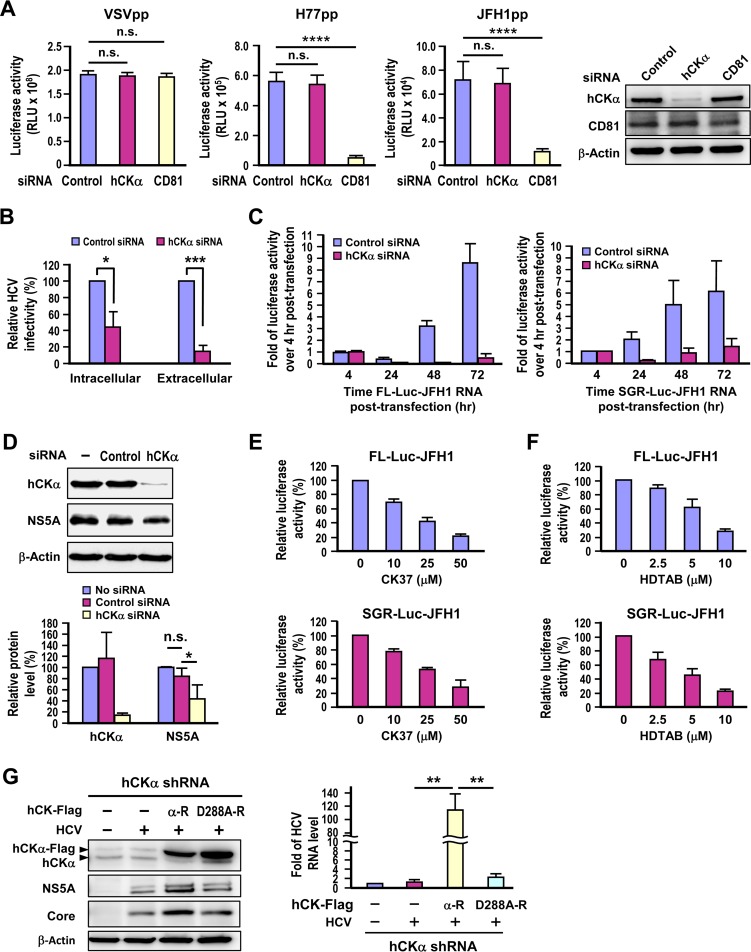
FIG 2.
Requirement of hCKα for HCV RNA replication. (A) Huh7 cells transfected with control or hCKα- or CD81-specific siRNAs for 48 h were separately infected with HCVpp generated from the genotype 1a H77 strain and genotype 2a JFH1 strain or with VSVpp, and luciferase activities were measured. In parallel, a set of cells harvested at 48 h post-siRNA transfection was analyzed by Western blotting; a representative data set is shown. (B) Huh7 cells transfected with control or hCKα-specific siRNAs were infected with HCV, and the infectivities of the extracellular and intracellular viruses were assessed. This analysis was performed by inoculating culture medium that contained extracellular virions and cell extracts that contained intracellular virions, respectively, into naive Huh7.5-1 cells. The infected cells were harvested, and isolated total RNA was analyzed for the HCV RNA level by RT-PCR. (C) Huh7 cells were transfected with FL-Luc-JFH1 RNA or SGR-Luc-JFH1 RNA, as indicated. HCV RNA replication was measured by determining the fold change in firefly luciferase activity at different time points over that determined at 4 h after RNA transfection. (D) Huh7 cells stably replicating genotype 1b Con1 strain SGR remained untransfected or were transfected with the indicated siRNAs, and cell lysates were analyzed by Western blotting (top panel). The level of the indicated proteins in different settings relative to that detected in siRNA-untreated SGR-replicating cells, which was arbitrarily designated 100%, was expressed as indicated (bottom panel). (E and F) FL-Luc-JFH1 or SGR-Luc-JFH1 RNA-transfected cells were treated with different concentrations of CK37 or HDTAB, as indicated, and the luciferase activity was measured and expressed as the percentage of that detected in cells that received vehicle treatment. (G) Huh7 cells constitutively expressing hCKα shRNA were transfected with control vector (−) or wild-type (α-R) or D288A (D288A-R) hCKα resistant to hCKα shRNA, and cells were left uninfected or infected with HCV. Duplicate sets of cells were analyzed by Western blotting for protein expression (left panel) and by RT-PCR for the viral RNA level (right panel). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.