ABSTRACT
Soluble forms of trimeric HIV-1 envelope glycoprotein (Env) have long been sought as immunogens and as reagents for analysis of Env structure and function. Isolation of trimers that mimic native Env, derived from diverse viruses, however, represents a major challenge. Thus far, the most promising native-like (NL) structures have been obtained by engineering trimer-stabilizing mutations, termed SOSIP, into truncated Env sequences. However, the abundances of NL trimeric conformers vary among Envs, necessitating purification by monoclonal antibodies (MAbs) like PGT145, which target specific epitopes. To surmount this inherent limitation, we developed an approach that uses lectin affinity chromatography, ion-exchange chromatography, hydrophobic-interaction chromatography (HIC), and size exclusion chromatography (SEC) to isolate NL trimers from nonnative Env species. We validated this method with SOSIP trimers from HIV-1 clades A and B. Analyses by SEC, blue native PAGE, SDS-PAGE, and dynamic light scattering indicated that the resulting material was homogeneous (>95% pure), fully cleaved, and of the appropriate molecular weight and size for SOSIP trimers. Negative-stain electron microscopy further demonstrated that our preparations were composed of NL trimeric structures. By hydrogen/deuterium-exchange mass spectrometry, these HIC-pure trimers exhibited structural organization consistent with NL trimers and inconsistent with profiles seen in nonnative Envs. Screened for antigenicity, some Envs, like BS208.b1 and KNH1144 T162A, did not present the glycan/quaternary structure-dependent epitope for PGT145 binding, suggesting that these SOSIPs would be challenging to isolate by existing MAb affinity methods. By selecting based on biochemical rather than antigenic properties, our method offers an epitope-independent alternative to MAbs for isolation of NL Env trimers.
IMPORTANCE The production and purification of diverse soluble Env trimers that maintain native-like (NL) structure present technical challenges that must be overcome in order to advance vaccine development and provide reagents for HIV research. Low levels of NL trimer expression amid heterogeneous Env conformers, even with the addition of stabilizing mutations, have presented a major challenge. In addition, it has been difficult to separate the NL trimers from these heterogeneous mixtures. While MAbs with specificity for quaternary NL trimer epitopes have provided one approach to purifying the desirable species, such methods are dependent on the Env displaying the proper epitope. In addition, MAb affinity chromatography can be expensive, the necessary MAb may be in limited supply, and large-scale purification may not be feasible. Our method based on biochemical separation techniques offers an epitope-independent approach to purification of NL trimers with general application to diverse Envs.
INTRODUCTION
The envelope glycoprotein (Env) on the virus surface is the sole target of HIV-1 neutralizing antibodies (NAbs). Many vaccine strategies involve the use of Env-based immunogens aimed at eliciting NAbs with broad cross-reactivity. Because the trimeric form of Env found on the surface of the virus mediates viral entry, and a major goal of vaccine design is to elicit antibodies that block this process, it is expected that immunogens must recapitulate the native structure of this functional Env (1–6). Diverse Env variants representing the most prevalent HIV-1 clades are currently under evaluation as potential immunogens.
Functional Env is a membrane-anchored trimer of extensively glycosylated heterodimers, composed of gp120 receptor-binding and gp41 membrane-spanning fusion subunits. Endogenous proteases cleave the gp160 precursor polypeptide into gp120 and gp41 subunits, which remain noncovalently associated as protomers in the native Env trimer. Current models of viral entry suggest that prior to receptor and coreceptor binding, Env adopts a “closed” prefusion conformation, in which conserved functional features such as receptor and coreceptor binding sites on the gp120 core and the V3 loop are “masked” or sterically inaccessible (7, 8). Dense clusters of N-linked glycans distributed across the surface of Env as well as on its flexible variable loops further shield conserved core features (9, 10). Despite the significant defenses that have evolved on Env to mask the conserved features, recent studies have demonstrated that broadly neutralizing antibodies (BNAbs) can target select epitopes distributed across much of the surface of the closed, prefusion form of Env (11).
As a type 1 fusion protein, in which the prefusion conformation exists as a high-energy, metastable state, Env is relatively prone to spontaneous transition to its postfusion state (12–14). It is believed that Env may have been additionally selected for a tendency to misfold, possibly as a mechanism of immune evasion, whereby immunodominant but nonnative, nonfunctional forms of Env are displayed on the virus surface alongside relatively few copies of functional, native trimer (15). As a consequence of these and other factors, early efforts to produce soluble, recombinant forms of Env glycoproteins were largely unsuccessful (16). First-generation uncleaved gp140 ectodomain constructs, which truncated the Env gene N-terminal to the transmembrane anchor, were found to lack native-like (NL) Env structural organization (16, 17). Such engineered proteins, derived from a range of Env sequences, in general appeared to adopt nonnative conformations in which the gp41 subunit degenerates to a highly stable, helical bundle state, resembling the postfusion conformation, while gp120 monomers remain loosely tethered or dissociated, with disordered V1/V2 and V3 loops (16, 17). As a consequence, uncleaved gp140s expose epitopes targeted by poorly neutralizing antibodies that are occluded on the native trimer (18). When used in vaccines, they are thus unlikely to elicit broadly neutralizing humoral responses that target the NL, closed prefusion trimer organization (18–21).
In order to overcome the limitations of this first generation of Env constructs, much of the recent focus of Env immunogen design has been on producing stabilized forms of trimeric Env that maintain the structural and antigenic features of the native closed prefusion conformation. A set of modifications to the Env sequence were identified that help reduce formation of nonnative Env proteins while enhancing formation of NL trimers (22–25). These modifications, when paired with selection of Env from specific viral variants that favor NL trimer formation, resulted in production of trimeric glycoprotein assemblies that closely resemble native Env trimers (26–30) and have led to breakthroughs in structure determination, which, in turn, identified additional stabilizing substitutions that lock the assembly in the closed, prefusion conformation (4, 31).
The most widespread approach to date for generating stabilized NL Env trimers employs SOSIP.v1 modifications (4, 26). In this approach, Env is truncated after residue 664 and thus lacks both the transmembrane and membrane-proximal domains of gp41; an Ile-to-Pro replacement is introduced in the first heptad repeat of the ectodomain of gp41 (gp41ecto), stabilizing the prefusion state; an engineered disulfide bridge links gp41ecto and gp120 subunits of each protomer to prevent their dissociation; and a hexa-arginine motif replaces the native cleavage sequence to enhance furin cleavage. Despite these modifications potentially altering some structural properties, this design and those based on it represent the most native-like reagent available for the study of Env using solution-based techniques. Purified, trimeric SOSIP.v1 Envs have been shown by numerous structural and biophysical techniques to faithfully mimic HIV-1 Env (27, 29, 32, 33) in its native, trimeric conformation (26). Furthermore, a SOSIP.v1 trimer engineered from the Env sequence of the transmitted form of the virus from an infant in the Nairobi Breastfeeding Clinical Trial (NBT), BG505.c2 T332N (26), effectively elicited potent neutralizing antibodies in rabbits and macaques (34). However, these NAbs lacked broad cross-reactivity—meaning that they failed to neutralize heterologous tier 2 and 3 viruses. It has therefore been proposed that elicitation of broadly neutralizing responses may require the use of polyvalent formulations of SOSIP Envs based on diverse viral variants to accomplish the hallmark goal of breadth and potency. Envs from different isolates, however, vary widely in their propensities to form recombinant NL trimers using the SOSIP design (35, 36). Consequently, numerous purification methods have been developed to isolate NL trimers from lower-yield Envs (26, 35, 36, 37, 43).
Positive selection of glycosylated proteins by affinity chromatography with lectins or glycan-dependent NAbs like 2G12 followed by gel filtration is a commonly employed method of SOSIP protein purification (26). While these steps provide some separation of Env species, such methods alone are not sufficient to isolate NL trimers from misfolded dimers and other nonnative conformations. A more stringent form of positive selection employs quaternary structure-dependent monoclonal antibodies (MAbs) PGT145 (35) and PGT151 (36). These approaches, based on selection of complexes that present the cognate epitope for the MAb employed, have proven effective and generalizable to SOSIP trimers designed from several isolates. An alternative, affinity-based method uses negative selection with antibodies such as F105 and GE1366 (37), which are directed toward epitopes that are exposed on dimeric and monomeric species but are occluded on the “closed” properly folded NL trimer. While such affinity methods have proven effective in purifying NL trimers designed from some isolates, they require the antigen to present specific protein and glycan epitopes that are not necessarily universal to isolates of interest. In most cases, the required antibodies are not widely available, and it also remains relatively cost-prohibitive to scale MAb purification as may be needed for vaccine production.
To circumvent the limitations of MAb affinity methods for SOSIP purification, we developed an approach that relies on biochemical rather than antigenic features of well-formed NL SOSIP trimers and applied it to the purification of SOSIP.v1 trimers based on diverse Env sequences. The method we describe is effective in isolating the NL trimer population even when it is a minority fraction of the total Env material. By making it possible to isolate a broad range of NL Env trimers and use them as reagents (e.g., for antigenicity assays, B cell sorting, and structural analyses), we anticipate that it will become possible to investigate the diversity of HIV-1 Env biology and Env-directed immune responses with greater rigor and scope than in the past. Since the approach we describe relies upon readily scalable biochemical methods, this and similar purification methods are expected to be amenable to production of large-scale preparations, which may be useful for vaccine production.
MATERIALS AND METHODS
Design of SOSIPs engineered from multiple HIV-1 envelope sequences.
The amino acid sequence of the canonical BG505.c2 SOSIP.v1 T332N was used as a template for design of new SOSIP.v1 constructs engineered from gp160 sequences of diverse HIV-1 isolates. Briefly, a disulfide bridge was introduced between cysteines at positions 501 and 605 (SOS) and an Ile-to-Pro mutation was introduced in the HR1 domain of gp41 (IP). We also modified the native proteolytic cleavage site 508REKR511 to a hexa-arginine motif, known to enhance cleavage by exogenous furin (26), and replaced the native leader sequence with that of the tissue plasminogen activator (tPa) to enhance secretion. New SOSIP genes were synthesized by Life Technologies with flanking PST1/NOT1 restriction sites and subsequently cloned into the pPPI4 mammalian expression vector (26). BG505.c2 and MG505.e1 SOSIP expression vectors were obtained from John Moore and colleagues. In their originally published forms, these genes encoded several added sequons for N-linked glycans in a conserved binding site for the monoclonal antibody 2G12. To better mimic the native antigenicity of the viral isolate, we reverted these glycan motifs to their wild-type sequences in the BG505 and MG505 SOSIPs using site-directed mutagenesis (Agilent).
Protein production and purification.
SOSIPs were produced by transient transfection with either 2× polyethylenimine (PEI) or Freestyle Max reagent (Life Technologies) in a suspension of 293F cells at between 0.8 and 1.2 million cells/ml. Cotransfection of furin in pcDNA.3 at a ratio of three SOSIPs to one furin ensured efficient proteolytic cleavage between gp120 and gp41 subunits during production. Six or 7 days after transfection, supernatants were cleared by centrifugation and filtered through a 0.2-μm vacuum filtration unit and supplemented with protease inhibitors (Roche) and sodium azide to prevent microbial growth. Supernatants were incubated with Galanthus nivalis lectin (GNL) coupled to agarose beads overnight at 4°C and washed with 20 mM Tris (pH 7.4), 1 mM EDTA, 1 mM EGTA, 0.02% azide, and 120 mM NaCl; glycoproteins were eluted with 7 to 10 column volumes of 1 M alpha methyl-mannopyrannoside dissolved in 20 mM Tris (pH 7.4), 1 mM EDTA, 1 mM EGTA, 0.02% azide, and 120 mM NaCl.
GNL eluates were concentrated using Amicon ultrafiltration units (nominal molecular mass cutoff of 100 kDa) and buffer exchanged to DEAE low-salt buffer (20 mM Tris [pH 8.0], 100 mM NaCl) before being loaded onto a 5-ml prepacked HiTrap DEAE anion-exchange column (GE Life Sciences). Following 10 min of isocratic flow in 100 mM NaCl, a gradient to 1 M NaCl was initiated and fractions were collected throughout. Alternatively, this step was done in batches while collecting the low-salt flowthrough, with a final elution of bound material with 1 NaCl.
The DEAE flowthrough was buffer exchanged into 2 M ammonium sulfate–0.1 M phosphate (pH 7.0) and loaded onto a ProPac HIC-10 column (Dionex). A linear gradient of 2 M to 0 M ammonium sulfate in 0.1 M phosphate (pH 7) over 90 min was sufficient to resolve trimers from dimers and monomers. Lastly, the early-eluting fractions (containing native-like trimers) were concentrated prior to being loaded onto a Superdex S200PG size exclusion chromatography (SEC) column in PBS (20 mM sodium phosphate [pH 7.4], 150 mM sodium chloride, 0.02% sodium azide). Peak fractions were collected and concentrated for downstream analyses of purity and antigenicity.
PGT145 pure BG505.c2 T332N SOSIP trimer was provided by John Moore and colleagues (original study described in reference 4). BG505.c2 T332N gp120 was produced and purified in-house by transient transfection of 293F cells, GNL affinity chromatography as previously described, and SEC on the Superdex S200PG column.
SDS-PAGE and BN-PAGE analyses.
SDS denaturing PAGE and blue native PAGE (BN-PAGE) analyses with precast gels (Novex) were performed to assess the oligomeric species present at each stage of purification. The addition of 0.1 M dithiothreitol (DTT) to denatured samples allowed us to ensure that furin cleavage between gp120 and gp41 subunits was complete in our SOSIPs. Typically, between 10 and 15 μg of protein was loaded per lane for BN-PAGE analysis and 5 μg per lane was loaded for SDS-PAGE analysis.
DLS.
Dynamic light scattering (DLS) measurements were performed on a Dynapro Nanostar (Wyatt Technologies). Trimer samples were diluted to 1 or 3 mg/ml in PBS and centrifuged at 15,000 × g for 20 min prior to loading of 8 μl into a low-volume quartz cuvette. The mean estimated radius, polydispersity, and molecular weight were generated from 40 acquisitions of 5 s at 20°C. Following these initial acquisitions, 10 μl of parrafin oil was gently added to the top of the sample prior to temperature scans from 30 to 80°C at a rate of 1°C/min to determine the onset of SOSIP melting and aggregation.
Negative-stain electron microscopy (NS-EM).
A 3-μl aliquot of purified SOSIP, diluted to 10 or 30 μg/ml in PBS, was applied for 60 s to glow-discharged C-Flat, 300-mesh Cu grids (Electron Microscopy Sciences) and stained for an additional 60 s using Nano-W (Nanoprobes). Data were collected using a FEI Tecnai T12 transmission electron microscope operating at 120 keV. Images were collected using a Gatan Ultrascan 4000 charge-coupled device (CCD) at a magnification of ×52,000 at 1.0-μm defocus, corresponding to a pixel size of 2.07 Å. Particles were selected using interactive particle picking in EMAN2.1 image processing suite. For each data set a phase-flipped, contrast transfer function (CTF)-corrected stack containing ∼20,000 particles was created and subjected to reference-free two-dimensional (2D) classification to generate ∼125 classes. Class averages containing multiple trimers within the particle box size were omitted, and a representative class average for each isolate was chosen for clarity.
BLI measurement of antibody-Env binding.
Biolayer interferometry (BLI) on an Octet red platform (ForteBio) was used to assess antigenic profiles of new SOSIPs purified by our method. Our measurements were performed in an assay buffer composed of phosphate-buffered saline supplemented with 1% bovine serum albumin (BSA), 0.03% Tween 20, and 0.01% azide at a shake speed of 1,000 rpm and 30°C. Tip regeneration between measurements was performed in 1 M glycine at pH 2.5. In a typical binding experiment, antibodies of interest and SOSIPs were diluted in assay buffer. We diluted antibodies to 8 μg/ml for loading onto dip-and-read biosensors coated in anti-human IgG. We diluted SOSIP analytes to a single concentration of 125 nM to assay association. Antigenicity screens were carried out as two independent replicates unless material was limiting (PVO.4).
The area, in nanometers per second, under each association/dissociation curve was calculated using Prism (GraphPad), and a heat map was generated based on these values to compare binding of SOSIPs to different antibodies. A smaller panel of antibodies was used for PVO.4, for which material was limiting. The area-under-the-curve heat map for PVO.4 binding is separated in Fig. 6 because association and dissociation times for these experiments differed from those used in the larger panel, making the inter-SOSIP comparison for this construct impossible at this time.
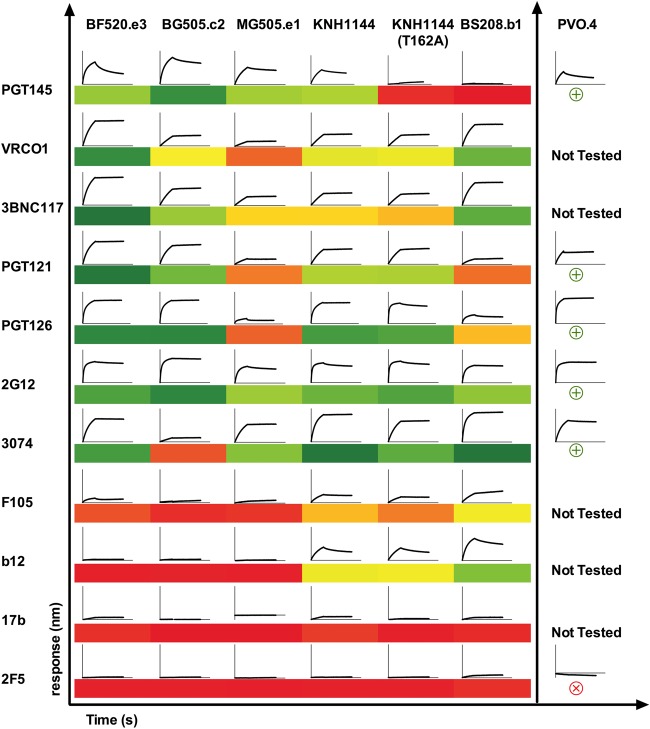
FIG 6.
Antigenicity of HIC pure trimers by biolayer interferometry (BLI). BLI analysis was used with a panel of HIV-1-specific neutralizing and nonneutralizing antibodies to assess antigenicity of trimers. For each antibody-Env interaction, we show a representative binding curve (association and dissociation phases) at 125 nM SOSIP. In the section on the left, SOSIPs for which enough material was available to assess binding to the entire panel are shown (BF520.e3, BG505.c2, MG505.e1, KNH1144 with or without N160, and BS208.b1). A heat map based on area under each binding curve, in which dark green indicates strong binding (>150 nm · s), yellow intermediate binding (<120 nm · s), and orange to red low or no binding (80 to 2 nm · s) on a continuous scale, is also shown for this panel to better compare these interactions. Note that BG505.c2, which lacks the N332 glycan, is still bound by BNAbs that target this epitope, likely due to promiscuous glycan recognition by PGT121 and PGT126 (42). For PVO.4, in the section on the right, material was limited and the association and dissociation times used for this earlier preparation were not consistent with antigenicity data from the other SOSIPs. We therefore summarize data from this Env separately and qualitatively, with a circled green plus sign and circled red cross sign indicating strong and low to no binding, respectively.
Hydrogen/deuterium-exchange mass spectrometry (HDX-MS).
Approximately 2 μg of protein for each time point was incubated at 22 to 25°C (room temperature) in a buffer containing deuterium (20 mM Na3PO4 [pH 7.4], 150 mM NaCl, 0.02% sodium azide, 1 mM EDTA, 85% D2O). The time course extended from 3 s to 20 h, and samples at each time point were incubated in duplicate. Samples were quenched by mixing 1:1 with 200 mM tris(2-chlorethyl) phosphate (TCEP)–0.02% formic acid for a final pH of 2.5 and flash frozen in liquid nitrogen. The zero time point was accomplished by premixing the deuterated buffer and quench solution before addition of protein. A fully deuterated control was also included in which denatured proteins (pretreated for 15 min at 85°C in 1 M DTT and 3 M guanidine hydrochloride) were deuterated for 1 h at 85°C.
Liquid chromatography-mass spectrometry (LC-MS) analysis was performed on a Waters Synapt G2-Si Q-TOF with an M class Aquity solvent delivery system. Samples were thawed briefly on ice and immediately injected onto a Waters HDX Manager kept at 1°C. Samples were passed over a 2.1- by 50-mm column filled with POROS-coupled pepsin at 150 μl/min and trapped onto a BEH 2.1- by 5-mm 1.7u C18 trap column. Peptides were resolved over a 1- by 100-mm BEH 1.7u C18 column (Waters) with a linear gradient of 5% solvent B to 50% solvent B in 10 min (solvent A, 0.1% fluoroacetic acid, 0.04% trifluoroacetic acid, and 5% acetonitrile; solvent B, 0.1% fluoroacetic acid and 100% acetonitrile). Source and desolvation temperatures were 70°C and 130°C, respectively, and the StepWave ion optics settings were adjusted to minimize loss of deuterium during ionization. Peptides were assigned from MSE measurements on undeuterated samples using ProteinLynx Global Server (PLGS; Waters Corp.) in combination with exact mass and known elution times from previous examinations of similar SOSIP constructs. Deuterium exchange kinetics were analyzed using HX-express v2. Percent exchange is reported relative to the values for the zero time point and fully deuterated samples. One set of data was lost during processing, resulting in only a single replicate time point for MG505, at 20 h. An internal peptide standard (Pro-Pro-Pro-Ile [PPPI]) was included in each sample to control for differences in ambient temperature at the time the experiments were performed.
RESULTS
Diverse Envs engineered as SOSIP.v1 vary widely in abundances of different conformers.
We examined six HIV-1 Env variants primarily from acute stages of infection, including both sexually and vertically transmitted viruses (Table 1); these Envs were from clades A and B and included a well-characterized, infant-derived Env, BG505.c2 (in this study in its wild-type form, lacking a glycan modification at position 332 [38], which was introduced into some SOSIP.v1 trimers to promote binding to glycan-specific MAbs like 2G12 [26]).
TABLE 1.
Summary of Env origins and traits
| Isolate | Origin | Transmission | Clade | Coreceptor | Infection | Tier | V1/V2 length | No. of predicted N-linked glycans per protomere | GenBank accession number | PubMed identifier |
|---|---|---|---|---|---|---|---|---|---|---|
| BG505.c2 | NBTa | MTCTb | A | R5 | Acute | 2 | 17/46 | 24 | ABA61516.1 | 16378985 |
| MG505.e1 | NBT | M-Fc | A | R5 | Chronic | NCf | 17/46 | 27 | ABA61511.1 | 16378985 |
| BF520.e3 | NBT | MTCT | A | R5 | Acute | 2 | 17/41 | 28 | KX168096 | 27345369 |
| BS208.b1 | NBT | MTCT | A | R5 | Acute | 2 | 13/42 | 22 | KX364401 | 16378985 |
| PVO.4 | Italy | M-Md | B | R5 | Acute | 2/3 | 29/48 | 32 | AAW64259.1 | 16051804 |
| KNH1144 | Discarded blood units from Kenyatta National Hospital | Unknown | A | R5 | Chronic | NC | 28/44 | 27 | AAN03143.1 | 12218394 |
NBT, Nairobi Breast Feeding Clinical Trial.
MTCT, mother-to-child-transmission.
M-F, male-to-female transmission.
M-M, male-to-male transmission.
Predicted using Los Alamos HIV sequence database N-GlycoSite software.
NC, not characterized.
When the six SOSIPs were purified by lectin affinity chromatography alone (sample analytical SEC shown in Fig. 1; analytical data for all SOSIPs shown in Fig. 2), BN-PAGE analysis revealed that all contained some fraction of trimer-sized species but that the relative abundance of trimers, dimers, and monomers—which, as glycoproteins, run heavier than expected on the native gel, at ∼700, 450, and 220 kDa, respectively—varied significantly across the panel (BN-PAGE left of NS-EM micrograph in Fig. 2); the amount of Env material that was aggregated and did not enter the native gel also varied among isolates but proved difficult to quantify by this method.
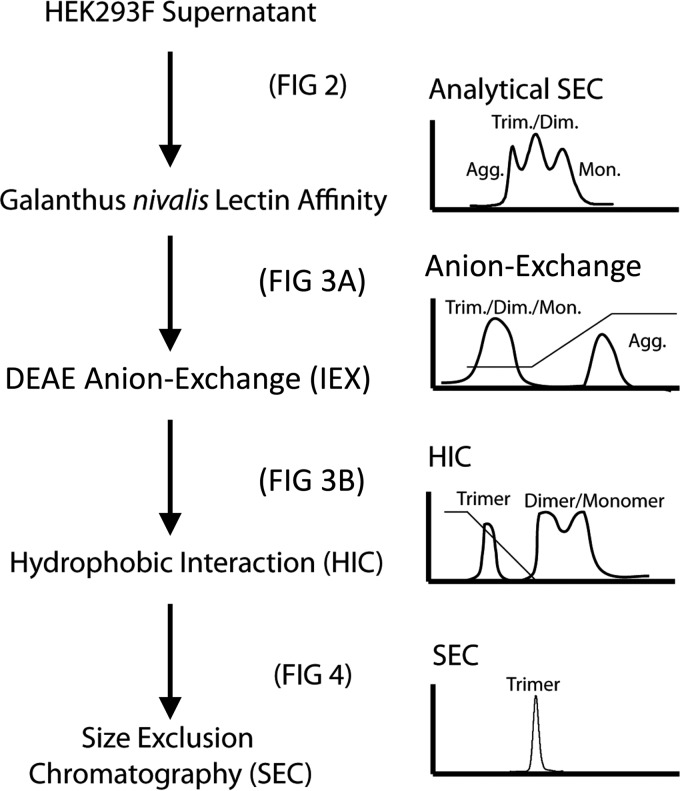
FIG 1.
Work flow for purification of SOSIP.v1 trimers. Stages of our preparative work flow are shown, with simulated chromatograms to the right of each purification step. The figures in which analyses of each step are shown are indicated.
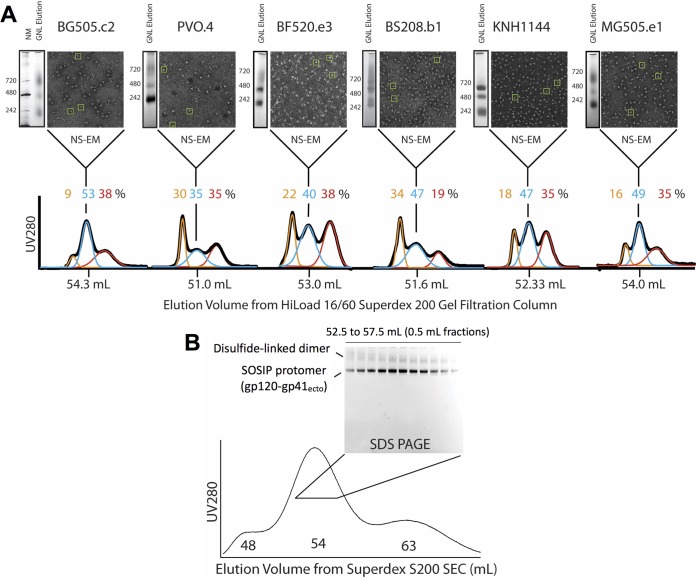
FIG 2.
(A) Characterization of trimer content after GNL affinity chromatography. We summarize our initial analysis of trimer content for each Env variant. To the left of each NS-EM micrograph, we show BN-PAGE analyses of eluted and concentrated protein after GNL affinity chromatography with molecular mass markers, in kilodaltons, indicated for each. These proteins were also analyzed by SEC, and analytical SEC chromatograms for proteins purified by GNL affinity chromatography are plotted, with elution volumes of each peak maximum indicated on the bottom x axis (black SEC trace). Each GNL profile was deconvoluted into three major peaks using Gaussian fits (orange, blue, and red), and the estimated percent that each peak contributes to the overall mixture is indicated in the corresponding color above the traces. The peak fraction of the central, NL trimer-containing peak for each variant was further analyzed by NS-EM. Representative particles that appear structurally consistent with NL trimer are boxed in each micrograph in green. (B) SEC and nonreducing SDS-PAGE analysis of GNL affinity chromatography-purified BG505.c2 SOSIP trimer. The SEC chromatogram profile for BG505.c2 GNL eluate is shown. Nonreducing SDS-PAGE was performed on fractions from 52.5 to 57.5 ml eluting in the central trimer peak to determine if any fraction was free from dimeric contaminants. gp120-gp41ecto SOSIP protomers, which run as gp140 after heating for SDS-PAGE, are labeled, as well as the dimer-contaminating material, which runs higher on the gel.
To quantify relative amounts of each Env species present after GNL affinity chromatography, we used size exclusion chromatography (SEC). In the SEC chromatograms shown in Fig. 2, each elution profile could be deconvolved into 3 major overlapping Gaussian peaks corresponding to different oligomeric Env species (fits are shown in orange, blue, and red). The earliest-eluting peak was composed of large aggregates eluting in the void volume of the SEC (orange trace in Fig. 2 chromatogram). This aggregated material contributed between 9 and 34% of the total mixture depending on the specific Env. The third peak (red trace) accounted for 19 to 38% of the concentrated protein eluting off SEC and was composed of a heterogeneous mixture of monomeric and dimeric Envs as assessed by BN-PAGE.
The middle peak in each chromatogram (blue trace in Fig. 2 chromatogram) contained the majority of NL trimer among all the fractions for each Env as assessed by nonreducing SDS-PAGE and confirmed by NS-EM. For the well-characterized BG505.c2 Env, by SEC, ∼53% of the total protein eluted in this peak, which had its A280 maximum in a position similar to that of purified NL glycosylated SOSIP trimers (14, 26, 35) but with a broader elution profile. In general, the other Envs had smaller amounts of protein that eluted in this middle peak, ranging from ∼35% in PVO.4 to 49% in MG505; all except MG505 were somewhat lower and significantly broader than for BG505.c2. For BG505.c2, on a nonreducing SDS-PAGE gel, the protein in this central peak was found to consist primarily of disulfide-bonded, cleaved gp120-gp41ecto SOSIP protomers that run close to 140 kDa (Fig. 2B). However, the gel analysis also revealed that the middle SEC peak contained significant, though variable, fractions of aberrantly disulfide bonded dimeric contaminants (Fig. 2B).
NS-EM enabled us to qualitatively assess trimeric structures present in the fraction of the central SEC peak with the maximum A280 absorbance for each Env. This peak trimer fraction was observed to contain significant amounts of recognizable NL trimers along with other nonnative Env species (representative trimer structures are boxed in green in micrographs in Fig. 2A). However, the ratio of NL trimer to nonnative-appearing structures varied among Envs, even in this peak SEC fraction, with BG505.c2, KNH1144, and MG505.e1 producing more particles consistent with NL SOSIP Envs relative to the others such as PVO.4 and BF520.e3, which showed fairly low abundances of recognizable NL trimers at this stage of purification.
DEAE weak anion-exchange chromatography removes aggregated Env.
Since GNL affinity chromatography alone yielded, at best, roughly half purified NL trimer, we next tested whether an additional ion-exchange step would help to remove aggregates and misfolded forms of Env (refer to Fig. 1 for purification scheme). Figure 3A shows the DEAE anion-exchange chromatogram (following GNL affinity purification) from our purification of BF520.e3 SOSIP trimer, which is representative of how all the Envs behaved. SEC analysis of the resulting post-ion-exchange step, BN- and SDS-PAGE analyses of the Env species, is also shown. We observed that the NL Env population flowed through the ion-exchange column under low-salt conditions (peak 1 in Fig. 3A that was collected during the 100 mM NaCl isocratic-flow stage of the chromatography run), while aggregated forms of Env and other contaminants were retained by the column and only eluted in high salt (peak 2 eluting in a gradient that increased the salt concentration to 1 M NaCl). The protein from each of these peaks was collected and subjected to analytical SEC to identify the oligomeric species that were present in the respective peaks. The DEAE flowthrough appeared to contain the Env species corresponding to trimer, dimer, and monomer (referenced to blue and red fitted traces in Fig. 2A SEC chromatograms). The SEC profile for the DEAE eluate (shown in red in Fig. 3A) matched the aggregated protein eluting in the void of the SEC column as observed from the GNL affinity SEC profiles shown in Fig. 2A. When the trimer peak fractions from the DEAE flowthrough (first peak in blue trace Fig. 3A) were analyzed by BN-PAGE, it was clear that this material was primarily trimeric and dimeric Env. The same analysis of the DEAE eluate showed an aggregated smear by BN-PAGE and extensive laddering by SDS-PAGE (Fig. 3A). We thus found that by adding a DEAE weak anion-exchange purification step after GNL affinity chromatography, we could separate NL trimer and smaller Env subassemblies from a more heterogeneous mixture of contaminants and aggregates produced by lectin affinity chromatography alone.
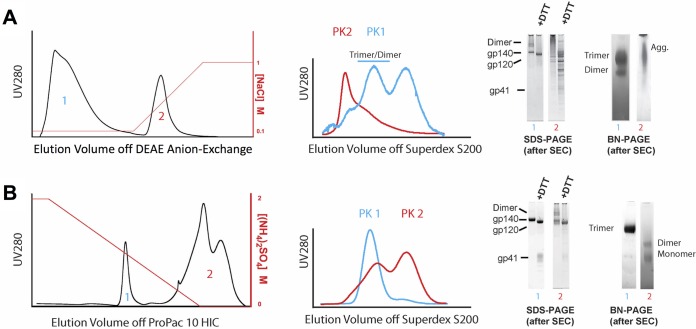
FIG 3.
Analyses of DEAE anion-exchange chromatography followed by hydrophobic-interaction chromatography (HIC) of GNL affinity chromatography-purified Envs. (A) A representative DEAE anion-exchange chromatogram from the purification of BF520.e3 is shown, with the increasing salt gradient indicated in red on the right axis. A fraction from each separated component (peaks [PK] 1 and 2) in this chromatogram was subjected to SEC for analysis. Overlaid chromatograms from this analysis are shown, with the SEC profile of peak 1 indicated in blue (low-salt flowthrough) and the SEC profile of peak 2 (high-salt eluate) indicated in red. Pooled fractions from these SEC chromatograms were further analyzed by reducing (+DTT) and nonreducing (−DTT) SDS-PAGE as well as BN-PAGE, shown in the gel panel to the right. (B) Trimer-containing material from peak 1 (blue) off the DEAE anion-exchange was next subjected to HIC. A representative chromatogram from the purification of BF520.e3 is shown, with the decreasing salt gradient indicated on the right y axis in red. Each separated component (peaks 1 and 2) was analyzed by SEC as in for panel A, and the overlaid chromatograms are shown. Fractions from these SEC chromatograms were further analyzed by SDS-PAGE and BN-PAGE, and these data are shown with numbers corresponding to each SEC chromatogram analyzed in the gel panel to the right.
HIC separates native-like Env trimers from other species.
Next we tested whether hydrophobic-interaction chromatography (HIC) applied after the DEAE ion-exchange step could further enrich NL trimer fractions and remove contaminating dimer and monomer fractions from the mixture of Env conformers. Figure 3B shows a representative HIC chromatogram from the purification of BF520.e3 SOSIP (second-to-last step shown in Fig. 1). Again, two primary populations were observed following separation by a salt gradient. The early peak (blue 1 in Fig. 3B) eluted with baseline resolution before the second eluting peak (red 2 in Fig. 3B), which came off the column only toward the end of the gradient. Each of these peaks was pooled and analytical SEC was run to determine the species that were present. Peak 1 from HIC (blue SEC trace in Fig. 3B), when passed over the SEC column, was eluted in a narrow, symmetrical profile at the volume expected for NL trimeric Env (∼54 ml). By BN-PAGE and reducing and nonreducing SDS-PAGE, this material appeared to consist of homogeneous, cleaved, NL trimeric Env (gel panel in Fig. 3B). When passed through the analytical SEC column, the material from peak 2 of the HIC step was eluted as two overlapping peaks (red trace in Fig. 3B). By BN and reducing and nonreducing SDS-PAGE, this HIC peak 2 material appeared to contain only dimeric and monomeric Envs (gel panel in Fig. 3B). Based on these analyses, it became clear why GNL affinity chromatography and SEC were insufficient to purify SOSIP trimers to homogeneity in the analysis in Fig. 2A. The dimeric species seen in the SEC trace of the second HIC peak clearly coeluted with the entirety of the true NL trimer.
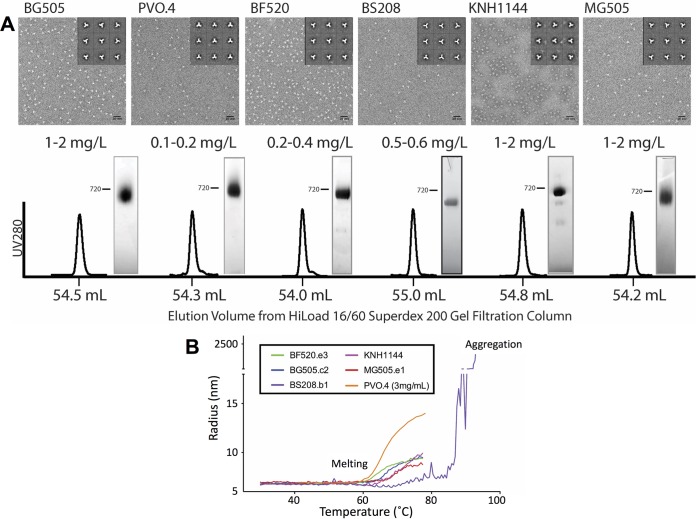
In contrast to the multiple, overlapping peaks that were observed for preparations that were processed only by lectin affinity chromatography (Fig. 2A), when the HIC-purified early-eluting peak for all six Envs was polished by SEC, we observed a single, well-resolved peak consistent in breadth and retention time with purified trimers for all Envs tested (Fig. 4A). These peaks were eluted consistently at ∼54 ml, which corresponds closely to the predicted molecular weight of SOSIP trimers (26, 35). BN-PAGE analysis also demonstrated that the purified SOSIP trimer preparations contained only the trimer populations, with little trace of possible nonnative subassemblies. By NS-EM, the predominance of NL trimer structures in the purified material was clearly evident (Fig. 4A). When the final material was run on reducing and nonreducing SDS-PAGE (representative data from BF520.e3 SOSIP are shown in the gel panel in Fig. 3B), it was also clear that all of the Env present was in the properly cleaved gp120-gp41 form, as the DTT reducing agent reverted the SOSIP disulfide-bonded gp120-gp41 to individual gp120 and gp41 subunits. These data confirmed that our method successfully separated cleaved, NL trimers from all other components of the mixture of Env assemblies produced by transient transfection in 293F cells.
FIG 4.
(A) Characterization of purified SOSIP trimers. SEC chromatograms from six HIC-purified trimers are shown, with peak fraction retention times indicated on the x axis. BN-PAGE analysis of the final trimer material is shown alongside these chromatograms, and an estimation of approximate yield in milligrams per liter of 293F transfection is indicated. NS-EM micrographs and trimeric class averages of this final material for each Env are also shown. (B) Dynamic light scattering (DLS) temperature scans of HIC-pure SOSIP trimers. Melting temperatures were assessed by measuring the change in particle radius over the course of a temperature scan from 30°C to 80°C (up to 90°C for BS208.b1). Melting was defined as a stable transition to between 9 and 15 nm from the closed-prefusion radius at baseline of ∼6.8 nm. Aggregation accompanied a more dramatic transition to 100- to 1,000-nm particle radii with multimodal polydispersity distributions (seen in the case of BS208.b1 at high temperatures).
Yields of NL trimers varied among isolates with BG505.c2, MG505.e1, and KNH1144 producing 1 to 2 mg of NL trimers per liter of transfection, which was higher than for other Envs. We found that PVO.4 produced as little as 0.1 mg/liter, necessitating significant scale-up to generate sufficient protein for our studies and even then limiting the antigenicity experiments we could undertake for this study. Nevertheless, even in this case, where the fraction of NL trimers was minor, the data show that the desirable subpopulation could be isolated and purified to homogeneity.
DLS confirms that the purified trimer material is monodisperse and of the expected size.
Dynamic light scattering (DLS) was used to measure the polydispersity of purified trimer preparations and the hydrodynamic radii of our HIC-purified SOSIP trimers in solution. This method can detect small levels of aggregation and contamination that may be difficult to assess by other methods such as BN-PAGE or SEC. All preparations showed a polydispersity of less than 7%, indicating that the samples were highly monodisperse and not significantly contaminated with other oligomeric species. Hydrodynamic radii measured by DLS ranged between 6.6 and 6.9 nm, consistent with trimer dimensions observed by NS-EM (Table 2).
TABLE 2.
Dynamic light scattering measurements of SOSIP.v1 trimer dimensions, purity, and thermal stability
| Env | Hydrodynamic radius (nm) | Polydispersity (%) | Molecular mass (kDa)a | Temp for onset of melting (°C) |
|---|---|---|---|---|
| BG505.c2 | 6.7 | 5.0 | 284.5 | 64.0 |
| PVO.4 | 6.6 | 6.2 | 281.6 | 60.3 (3 mg/ml) |
| BF520.e3 | 6.7 | 4.3 | 292.8 | 63.3 |
| BS208.b1 | 6.5 | 3.4 | 268.1 | 68.4 (temp for onset of aggregation, 84.7) |
| KNH1144 | 6.7 | 4.7 | 287.0 | 62.6 |
| KNH1144 A162T | 6.8 | 3.9 | 300.0 | 65.8 |
| MG505.e1 | 6.8 | 2.7 | 301.95 | 65.3 |
Molar mass derived from DLS estimates of size, density, and particle conformation.
DLS was also used to measure the thermal stability of the various SOSIP trimers. In this case, DLS measurements were gathered as the temperature of the thermostated sample cuvette was gradually increased from 20 to 80°C. All trimers exhibited a stable baseline and single transition onset between 60 and 70°C, consistent with the thermal stability seen in previous studies of SOSIP trimers (26, 35, 36) (representative melting curves are shown in Fig. 4B). Following the temperature-induced transition, the majority of the samples adopted some larger form of the trimer that subsequently appeared to aggregate, as indicated by the dramatic increase in apparent particle dimensions above ∼80°C (data not shown). The BS208 SOSIP.v1 trimer showed a different phenotype in that it did not exhibit a clear onset of melting or transition prior to aggregation at extremely high temperatures (∼85°C) (purple trace in Fig. 4B).
HDX-MS to probe local structural order and conformational state.
We used HDX-MS to examine whether the HIC-purified SOSIP trimers exhibited exchange profiles and local structural organization consistent with NL trimers. The deuterium uptake profiles for peptides covering N/C gp120 termini and V2, V3, and gp41 segments, which are diagnostic of NL trimer formation (uptake profiles shown in Fig. 5) (14, 17), were in excellent agreement with previously reported data from our group for PGT145-purified BG505.c2 (T332N mutant) (4). Only subtle differences between PGT145 pure and HIC-purified material were apparent, which likely originate from the two data sets having been collected at different times under slightly different ambient conditions, an effect that is apparent in the PPPI internal standard, a recombinant tetrapeptide included to probe for such differences (Fig. 5). In contrast, HDX-MS data for monomeric gp120 from BG505.c2 T332N showed a considerably less ordered profile based upon these diagnostic peptides. The monomeric gp120 behaved in a manner consistent with the V1/V2 and V3 lacking stabilizing interactions, and the N/C termini likewise were highly dynamic in the absence of interactions with a gp41 subunit. Among the trimers, some individual segments did show isolate-specific differences such as in the case of BF520.e3, which appears to present a more dynamic V2 loop relative to the other SOSIP trimers. The rest of the diagnostic peptides for BF520.e3 are in excellent agreement with the other trimers and indicate that the differences are primarily localized to the V1/V2 apex.
FIG 5.
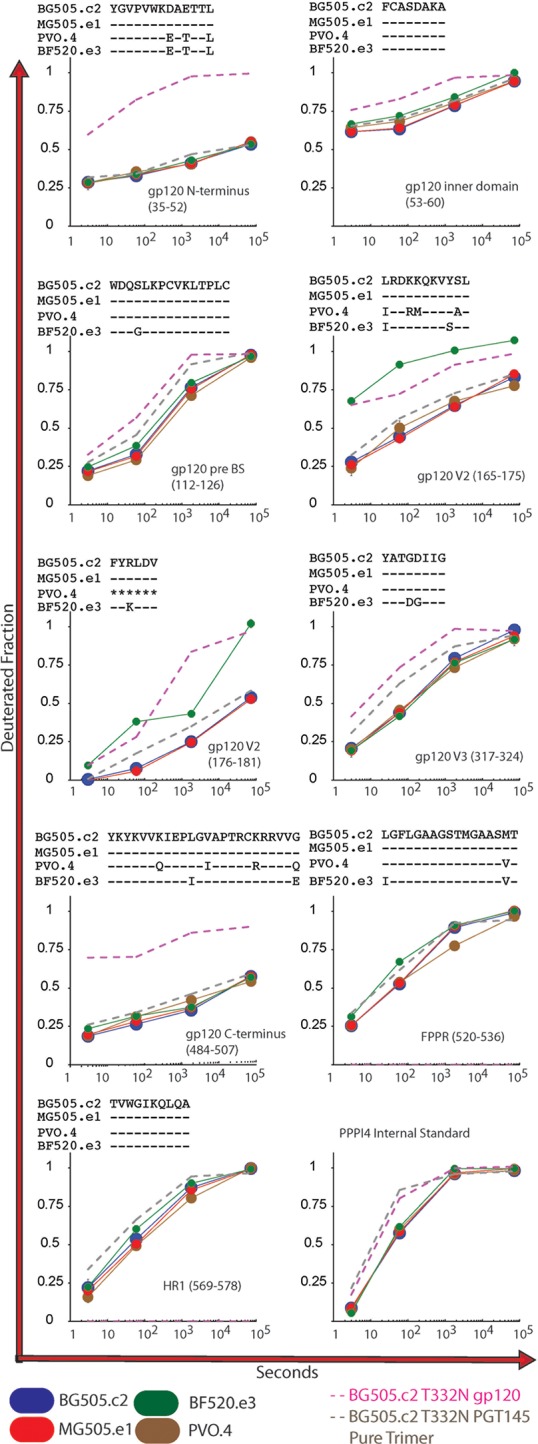
HDX-MS of HIC pure trimers. We examined a subset of our HIC-pure SOSIP trimers using hydrogen/deuterium-exchange mass spectrometry (HDX-MS). Deuterium-uptake profiles (BG505.c2 in blue, MG505.e1 in red, PVO.4 in brown, and BF520.e3 in green) are shown for N-terminal peptides of the gp120 inner domain, pre-bridging sheet segment (pre-BS), V2, V3, C terminus, fusion peptide-proximal region (FPPR), and first heptad repeat (HR1) domains. The deuterated fraction is plotted for each Env and time point in duplicate (except for MG505.e1 at the 20-h time point, for which only one data set was available). For reference, we also included data for peptides from monomeric gp120 (pink dashed line) and PGT145-purified BG505 T332N trimer (gray dashed line). Each reference data set (monomer and trimer) was collected in experiments separately from the HIC-pure trimers, under slightly different conditions, and only single time points are included to establish patterns for native-like (SOSIP) and nonnative (gp120) Env.
Monoclonal antibody recognition of HIC-purified SOSIP trimers.
Using biolayer interferometry to measure binding of the purified trimers to IgG captured on the Octet sensors, we observed diverse antigenic profiles for our HIC-pure SOSIP trimers against a panel of HIV-specific MAbs with known epitopes (Fig. 6). BG505.c2, MG05.e1, PVO.4, and KNH1144 bound robustly (areas under single binding curves ranging from 153 to 170 nm · s) to V1/V2 quaternary specific antibody PGT145. Binding was lower (7 to 19 nm · s) for BS208.b1—an Env with notably short loops at its apex—and for KNH1144 (T162A), a mutant we purified by the same method lacking a critical glycan for PGT145 binding. BF520.e3 bound PGT145 with strong and rapid association and relatively fast dissociation, likely resulting in weak affinity.
HIC-purified SOSIP trimers also showed low binding (<40 nm · s) over short periods to F105, b12, and 17b—antibodies with epitopes that are difficult to access and bind in the NL trimeric conformation (10) of most Env isolates studied to date. Notable exceptions included KNH1144 and BS208.b1, which both bound to b12 and to some extent to F105. As a negative control for Octet BLI-monitored antibody binding we included 2F5, an membrane-proximal external region (MPER) antibody targeting an epitope not present in SOSIP trimers. None of the HIC-purified trimers showed appreciable binding to this negative-control IgG.
It is important to note that timings were consistent across this panel for association and dissociation for all variants except PVO.4, for which limited material prevented a repeat experiment with timing identical to that of the other curves. For this reason, we have offset the PVO.4 binding data in Fig. 6 and compared curves internally to only other PVO.4-Ab interactions. We also note that a single concentration was not sufficient to rigorously derive kinetic parameters for these interactions. While we can conclude semiquantitatively that certain interactions are high-level binding or low-level binding and attribute these observations to either rapid or slow association and dissociation phases, we cannot definitively compare similar interactions using this screen.
DISCUSSION
We present a scalable approach for purifying native-like (NL) SOSIP trimers that does not require HIV-specific monoclonal antibodies. The combination of lectin affinity, anion-exchange chromatography, hydrophobic-interaction chromatography, and SEC was sufficient to separate NL trimers from nonnative conformations in preparations of SOSIPs engineered from multiple diverse Envs, many derived from acute and transmitted/founder viruses. Based on the level of purity obtained for these Env SOSIPs, we propose that this approach could be used in the place of or perhaps in addition to existing affinity methods for the purification of NL trimeric immunogens, which have become promising candidates for HIV-1 vaccines. And because other modified forms of engineered, soluble trimer such as “NFL” or “NFL TD” (39) and “single-chain” (40) are likely to display similar biochemical profiles with the hydrophilic glycan shield presented on the exterior of natively structured assemblies, we anticipate that the purification approach we describe will also be effective in isolating the NL trimeric population from these recombinant systems.
In addition to applications in immunogen production, our method enables study of variant-specific Env structural features, which may underlie differences in antigenicity and function. Our initial structural analysis by HDX-MS and NS-EM of this panel of Env SOSIP trimers demonstrated that while our purified trimers retain the closed, prefusion NL Env conformation, some specific structural differences were identifiable, and some of these appear to correlate with antigenic differences. By HDX-MS, the BF520.e3 trimer shared similar structural order in regions probed by most diagnostic peptides; however, in peptides from V2, spanning 165 to 181, significant deuterium exchange was observed, suggesting that those stretches of V2 may be more dynamic and exposed in this trimer than in other Envs (Fig. 5). This property was accompanied by a distinct mode of interaction with the quaternary specific BNAb PGT145, an antibody that does not neutralize the corresponding BF520.e3 pseudovirus but does bind strongly to transiently expressed Env on the surface of cells. The epitope for PGT145 is located proximal to the V2 segment, where deuterium uptake was rapid in this BF520.e3 trimer and we observed rapid association—comparable to those observed for BG505.c2 and MG505.e1. The dissociation, however, was considerably faster, suggesting that a highly specific but transient interaction was taking place, which may be attributable to a difference in Env structural dynamics in the transition of this isolate from “closed” to “open” prefusion conformations.
Characterization of trimer organization by NS-EM also revealed apparent structural differences of one Env variant, BS208.b1, which displayed trimeric lobes that were more slender and lacked the curved, blade-like propeller morphology seen in the other trimers (Fig. 4A). We hypothesize that this difference could be related to the differences in V1/V2 interactions at the apex of the BS208.b1 trimer relative to Envs such as BG505.c2 (Table 1). BS208.b1 has relatively short V1/V2 loops, with fewer potential N-linked glycosylation sites than the other trimers we examined; for example, the V1 length in BS208.b1 is 13, versus 17 in BG505.c2, and the V2 length is 42, versus 46. As in the case of the BF520.e3 SOSIP, BS208.b1 trimers interact weakly with the antibody PGT145, suggesting that a change in organization and presentation of the antibody's epitope at the apex exists in BS208.b1 trimers, despite conservation of the N160 and N156 glycosylation sites that are often targeted by this class of antibodies (Fig. 6). The distinct morphology and variable loop organization may also relate to the robustness of this Env to thermal perturbation, where the DLS-monitored thermal melts indicated that BS208.b1 trimers show a delayed onset of melting and aggregation (Fig. 4B).
Because BF520.e3, BS208.b1, and KNH1144 T162A SOSIPs have structural features that ablate or perturb high-affinity interactions with the quaternary specific antibody PGT145, we propose that these and similar Envs would be poor candidates for purification by current PGT145-based procedures (35). In such cases, we would predict very low yields due to low affinity or negligible binding of these Envs to the PGT145 affinity column. We further propose that an inability to purify by PGT145 is not necessarily globally diagnostic of poor or deficient trimer formation. Clearly, native and functional envelope trimers are produced for these variants given that the corresponding pseudotyped viruses are infectious (38, 41). Our results suggest that the SOSIP-modified, soluble forms of these Env variants retain an NL trimeric structure. Our approach thus appears to have significant and broad utility in isolating NL trimers from antigenically and phenotypically diverse Envs, including those that would be challenging to purify using PGT145 affinity approaches.
In summary, we have developed a new method for NL Env trimer purification that separates Env conformers based upon their general biochemical/biophysical properties, such as surface charge and exposure of hydrophobic patches. This procedure produces highly pure NL Env and avoids the need for using Env-specific MAbs. With this method, we have demonstrated the application to diverse Env trimers that retain unique structural properties such as localized structural differences that are detectable by HDX-MS. These structural differences in specific regions within the context of overall NL trimers showed reasonable correlations with antibody binding to those specific sites and epitopes. With the ability to isolate diverse NL Env trimers, we anticipate that it will be possible to expand our understanding of structural, phenotypic variation in HIV-1 as well as to provide reagents for immunogen production.
ACKNOWLEDGMENTS
We thank John Moore and colleagues for providing the BG505.c2 T332N and nonmutated MG505.e1 SOSIP constructs, which were reverted for this study. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program: CD4-IgG2 was from Progenics Pharmaceuticals, Inc.; HIV-1 gp120 monoclonal antibody 17b was from James E. Robinson; VRC01 was from John Mascola; F105 was from Marshall Posner and Lisa Cavacini; b12 was from Dennis Burton and Carlos Barbas; 3074 was from Susan Zolla-Pazner; 2G12 and 2F5 were from Herman Katinger; and PGT121, PGT126, and PGT145 were from IAVI.
REFERENCES
- 1.Gao F, Weaver EA, Lu Z, Li Y, Liao H-X, Ma B, Alam SM, Scearce RM, Sutherland LL, Yu J-S, Decker JM, Shaw GM, Montefiori DC, Korber BT, Hahn BH, Haynes BF. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J Virol 79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beddows S, Schülke N, Kirschner M, Barnes K, Franti M, Michael E, Ketas T, Sanders RW, Maddon PJ, Olson WC, Moore JP. 2005. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 79:8812–8827. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng C, Pancera M, Bossert A, Schmidt SD, Chen RE, Chen X, Druz A, Narpala S, Doria-Rose NA, McDermott AB, Kwong PD, Mascola JR. 2016. Immunogenicity of a prefusion HIV-1 envelope trimer in complex with a quaternary-structure-specific antibody. J Virol 90:2740–2755. doi: 10.1128/JVI.02380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien J-P, van den Kerkhof TLGM, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. 2015. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell 163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Wyatt R, Sodroski J. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol 75:1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sliepen K, Ozorowski G, Burger JA, van Montfort T, Stunnenberg M, LaBranche C, Montefiori DC, Moore JP, Ward AB, Sanders RW. 2015. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology 12:82. doi: 10.1186/s12977-015-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, Hedestam GBK, Wyatt RT. 2014. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A 111:E738–E747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang Z-Y, Zhang M-Y, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart-Jones GBE, Soto C, Lemmin T, Chuang G-Y, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens A-J, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee C-CD, Wu C-Y, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong C-H, Mascola JR, Kwong PD. 2016. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165:813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 11.Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien J-P, Lee JH, van Montfort T, de Taeye SW, Connors M, Burton DR, Wilson IA, Klasse P-J, Ward AB, Moore JP, Sanders RW. 2015. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog 11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert DM, Kim PS. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem 70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 13.Park HE, Gruenke JA, White JM. 2003. Leash in the groove mechanism of membrane fusion. Nat Struct Mol Biol 10:1048–1053. doi: 10.1038/nsb1012. [DOI] [PubMed] [Google Scholar]
- 14.Guttman M, Garcia NK, Cupo A, Matsui T, Julien J-P, Sanders RW, Wilson IA, Moore JP, Lee KK. 2014. CD4-induced activation in a soluble HIV-1 Env trimer. Structure 22:974–984. doi: 10.1016/j.str.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sattentau QJ. 2013. Envelope glycoprotein trimers as HIV-1 vaccine immunogens. Vaccines 1:497–512. doi: 10.3390/vaccines1040497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, Korzun J, Derking R, van Montfort T, Julien J-P, Wilson IA, Klasse PJ, Ward AB, Moore JP. 2013. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci U S A 110:18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman M, Lee KK. 2013. A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J Virol 87:11462–11475. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasmeen A, Ringe R, Derking R, Cupo A, Julien J-P, Burton DR, Ward AB, Wilson IA, Sanders RW, Moore JP, Klasse PJ. 2014. Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology 11:41. doi: 10.1186/1742-4690-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guttman M, Cupo A, Julien J-P, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorman J, Soto C, Yang MM, Davenport TM, Guttman M, Bailer RT, Chambers M, Chuang G-Y, DeKosky BJ, Doria-Rose NA, Druz A, Ernandes MJ, Georgiev IS, Jarosinski MC, Joyce MG, Lemmin TM, Leung S, Louder MK, McDaniel JR, Narpala S, Pancera M, Stuckey J, Wu X, Yang Y, Zhang B, Zhou T, Program NCS, Mullikin JC, Baxa U, Georgiou G, McDermott AB, Bonsignori M, Haynes BF, Moore PL, Morris L, Lee KK, Shapiro L, Mascola JR, Kwong PD. 2016. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol 23:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Julien J-P, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schülke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PWHI, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol 76:7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, Travis B, Kuhmann S, Burton DR, Hu S-L, Olson WC, Moore JP. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 76:2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders RW, Schiffner L, Master A, Kajumo F, Guo Y, Dragic T, Moore JP, Binley JM. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J Virol 74:5091–5100. doi: 10.1128/JVI.74.11.5091-5100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, Golabek M, de los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyumkis D, Julien J-P, de Val N, Cupo A, Potter CS, Klasse P-J, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse P-J, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang G-Y, Ofek G, Stewart-Jones GBE, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang G-Y, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GBE, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schön A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, McDermott AB, Mascola JR, Kwong PD. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JLS, Wilson IA, Olson WC, Moore JP, Subramaniam S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci U S A 108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramírez M, Back JW, Koff W, Julien J-P, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, de los Reyes K, Ketas TJ, Julien J-P, Burton DR, Wilson IA, Sanders RW, Klasse PJ, Ward AB, Moore JP. 2015. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol 89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Julien J-P, Lee JH, Ozorowski G, Hua Y, de la Peña AT, de Taeye SW, Nieusma T, Cupo A, Yasmeen A, Golabek M, Pugach P, Klasse PJ, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci U S A 112:11947–11952. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. 2015. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog 11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SMJ, Overbaugh J. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol 80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. 2016. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol 90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, Schön A, Galen JV, Chuang G-Y, Gorman J, Harned A, Pancera M, Stewart-Jones GBE, Cheng C, Freire E, McDermott AB, Mascola JR, Kwong PD. 2015. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol 89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonich CA, Williams KL, Verkerke HP, Williams JA, Nduati R, Lee KK, Overbaugh J. 2016. HIV-1 neutralizing antibodies with limited hypermutation from an infant. Cell 166:77–87. doi: 10.1016/j.cell.2016.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien J-P, Menis S, Wickramasinghe L, Seaman MS, Schief WR, Wilson IA, Poignard P, Burton DR. 2014. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med 6:236ra63. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.AlSalmi W, Mahalingam M, Ananthaswamy N, Hamlin C, Flores D, Gao G, Rao VB. 2015. A new approach to produce HIV-1 envelope trimers. J Biol Chem 290:19780–19795. doi: 10.1074/jbc.M115.656611. [DOI] [PMC free article] [PubMed] [Google Scholar]