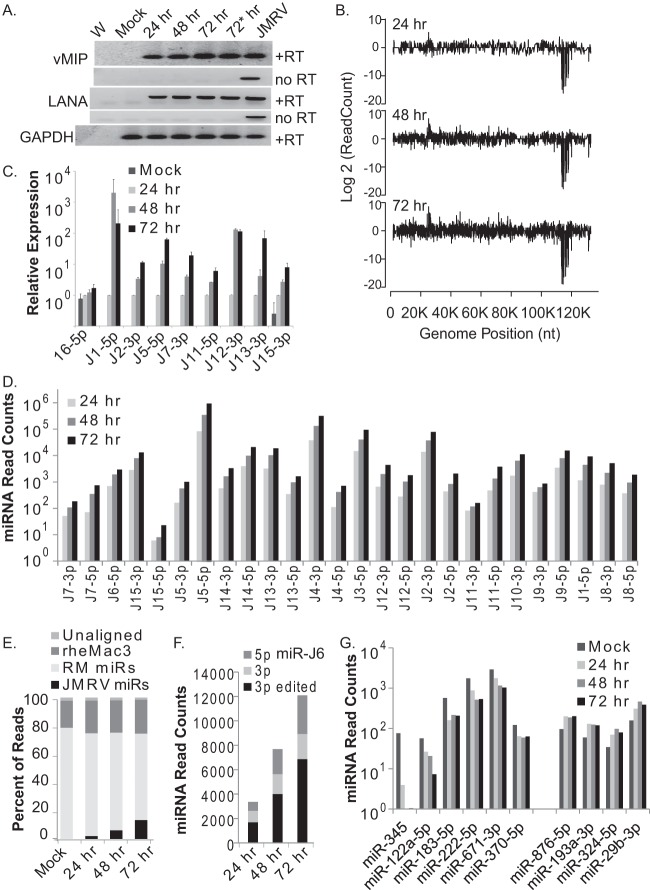
FIG 3.
miRNA expression kinetics during JMRV lytic infection. (A) RT-PCR analysis of vMIP, LANA, and cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in uninfected primary RFs (mock) or RFs infected with JMRV for the times indicated (MOI of 0.85 for the 24-, 48-, and 72-h samples and MOI of 2 for the 72-h* sample). PCR products were analyzed on a 1% TAE agarose gel and stained with ethidium bromide for visualization. +RT and no RT indicate analyses with or without reverse transcriptase. W, nuclease-free water (negative control); JMRV, genomic DNA from JMRV-infected cells (positive control). (B) Small-RNA sequencing analysis of JMRV-infected RFs at 24, 48, and 72 h postinfection. Shown are reads aligning across the JMRV genome. As described in the legend of Fig. 2, reads aligning to the plus strand are shown as positive values, while reads aligning to the minus strand are shown as negative values on the y axis. (C) TaqMan qRT-PCR analysis of cellular miR-16-5p and select JMRV miRNAs in uninfected (mock) or JMRV-infected RFs (24, 48, or 72 h postinfection). Expression values are normalized to values for RNU6B and are reported relative to miRNA levels at 24 h postinfection (set to “1”). (D) Normalized read counts for each JMRV miRNA (5p and 3p strands) at 24 h, 48 h, or 72 h postinfection as determined by deep sequencing. miRNAs are ordered from left to right based on their position in the JMRV genome. No reads were detected for miR-J10-5p. miR-J6-3p reads are shown separately in panel F. (E) Distribution of mapped reads for each of the four small-RNA sequencing libraries (mock- and JMRV-infected samples). (F) Normalized read counts for the edited form or nonedited form of miR-J6-3p as well as the 5p arm detected in each JMRV-infected sample. (G) Cellular miRNAs significantly altered (P value of <0.05 and FDR of <0.05) during JMRV lytic infection. Normalized miRNA read counts for six downregulated and four upregulated cellular miRNAs are shown.